A metatranscriptomic exploration of fungal and bacterial contributions to allochthonous leaf litter decomposition in the streambed
- PMID: 40297467
- PMCID: PMC12036579
- DOI: 10.7717/peerj.19120
A metatranscriptomic exploration of fungal and bacterial contributions to allochthonous leaf litter decomposition in the streambed
Abstract
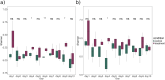
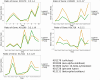
The decomposition of organic matter is essential for sustaining the health of freshwater ecosystems by enabling nutrient recycling, sustaining food webs, and shaping habitat conditions, which collectively enhance ecosystem resilience and productivity. Bacteria and fungi play a crucial role in this process by breaking down coarse particulate organic matter (CPOM), such as leaf litter, into nutrients available for other organisms. However, the specific contribution of bacteria and their functional interactions with fungi in freshwater sediments have yet to be thoroughly explored. In the following study, we enriched organic matter through the addition of alder (Alnus glutinosa) leaves into artificial stream channels (AquaFlow mesocosms). We then investigated enzyme expression, metabolic pathways, and community composition of fungi and bacteria involved in the degradation of CPOM through metatranscriptomics and amplicon sequencing. Enzymes involved in the degradation of lignin, cellulose, and hemicellulose were selectively upregulated with increased organic matter. Analysis of ITS and 16S rRNA gene sequences revealed that during decomposition, fungal communities were predominantly composed of Basidiomycota and Ascomycota, while bacterial communities were largely dominated by Pseudomonadota and Bacteroidota. The similar gene expression patterns of CPOM degradation related enzymes observed between bacteria and fungi indicate potential functional interaction between these microbial groups. This correlation in enzyme expression may indicate that bacteria and fungi are jointly involved in the breakdown of coarse particulate organic matter, potentially through mutualistic interaction. This study uncovers the specific enzymatic activities of bacteria and fungi and the importance of microbial interactions in organic matter decomposition, revealing their central role in facilitating nutrient cycling and maintaining the ecological health and stability of freshwater ecosystems.
Keywords: Coarse perticulate organic matter; Enzyme activity; Freshwater ecosystem health; Metatranscriptomics; Microbial interactions; Sequencing.
© 2025 Deep et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
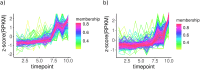


References
-
- Abarenkov K, Nilsson RH, Larsson KH, Taylor AFS, May TW, Frøslev TG, Pawlowska J, Lindahl B, Põldmaa K, Truong C, Vu D, Hosoya T, Niskanen T, Piirmann T, Ivanov F, Zirk A, Peterson M, Cheeke TE, Ishigami Y, Jansson AT, Jeppesen TS, Kristiansson E, Mikryukov V, Miller JT, Oono R, Ossandon FJ, Paupério J, Saar I, Schigel D, Suija A, Tedersoo L, Kõljalg U. The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: sequences, taxa and classifications reconsidered. Nucleic Acids Research. 2024;52(D1):D791–D797. doi: 10.1093/nar/gkad1039. - DOI - PMC - PubMed
-
- Abdel-Raheem A, Shearer C. Extracellular enzyme production by freshwater ascomycetes. Fungal Diversity. 2002;11:1–19.
-
- Abril M, Menéndez M, Ferreira V. Decomposition of leaf litter mixtures in streams: effects of component litter species and current velocity. Aquatic Sciences. 2021;83 doi: 10.1007/s00027-021-00810-x. 54. - DOI
-
- Apprill A, McNally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microbial Ecology. 2015;75(2):129–137. doi: 10.3354/ame01753. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

