The role of exosomal lncRNAs in mediating apoptosis and inflammation in UV-induced skin photoaging
- PMID: 40297520
- PMCID: PMC12034729
- DOI: 10.3389/fcell.2025.1538197
The role of exosomal lncRNAs in mediating apoptosis and inflammation in UV-induced skin photoaging
Abstract
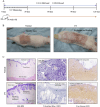
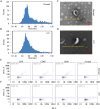
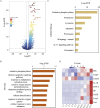
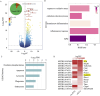
The skin, as the body's largest organ, functions as a vital barrier against environmental insults. Chronic exposure to ultraviolet (UV) radiation significantly contributes to premature aging, or photoaging, which leads to DNA damage and disrupts repair mechanisms. Exosomes, which are small extracellular vesicles, play a key role in cell-to-cell communication and might help mitigate the effects of photoaging by transporting bioactive molecules to skin cells. Long non-coding RNAs (lncRNAs) are increasingly recognized for their regulatory roles in the photoaging process, influencing stress responses and DNA repair; however, their involvement in exosomes in the context of skin aging is not yet well understood. In this study, we developed a photoaging model using SD rats subjected to UVA and UVB irradiation, which led to significant changes in the dermis such as increased dryness, wrinkles, pigmentation, and vascular alterations. Histological evaluations showed uneven thickening of the epidermis, degradation of collagen and elastic fibers, and cellular infiltration. Exosomes isolated from the dermal tissues exposed to UV radiation displayed altered size distributions. Transcriptomic analyses of the UV-treated rats identified 2,332 lncRNAs and 5,906 mRNAs that were differentially expressed, revealing significant involvement in pathways related to oxidative stress, apoptosis, and cellular stress responses. A cis-regulatory analysis identified 1,327 essential interactions between lncRNAs and mRNAs, highlighting their role in controlling inflammation and apoptosis. Importantly, both IL-1B and GADD45B levels were significantly increased in the exosomes and UV-challenged HaCaT cells, indicating their crucial roles in responding to UV-induced stress. This study highlights the significant role of exosomal lncRNAs in managing cellular reactions to UV-induced stress, impacting regulatory pathways associated with apoptosis, inflammation, and oxidative stress. These insights pave the way for the development of lncRNA-focused therapeutic approaches to address UV-induced skin damage.
Keywords: cis-regulatory; exosome; lncRNA; rat; skin photoaging.
Copyright © 2025 Li, Lin, Zhou, Guo, Lin and Ji.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Abel F., Murke F., Gaida M., Garnier N., Ochsenfarth C., Theiss C., et al. (2020). Extracellular vesicles isolated from patients undergoing remote ischemic preconditioning decrease hypoxia-evoked apoptosis of cardiomyoblasts after isoflurane but not propofol exposure. PLoS One 15, e0228948. 10.1371/journal.pone.0228948 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

