This is a preprint.
Glucuronidation Metabolomic Fingerprinting to Map Host-Microbe Metabolism
- PMID: 40297700
- PMCID: PMC12036448
- DOI: 10.21203/rs.3.rs-6321321/v1
Glucuronidation Metabolomic Fingerprinting to Map Host-Microbe Metabolism
Abstract
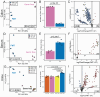
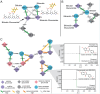
Glucuronidation is an important detoxification pathway that operates in balance with gastrointestinal microbial β-glucuronidase (GUS) enzymes that can regenerate active metabolites from their glucuronidated forms. Although significant progress has been made in characterizing GUS enzymes, methods to comprehensively define the glucuronidome - the collection of glucuronidated metabolites - remain limited. In this study we employed pattern-filtering data science approaches alongside untargeted LC-MS/MS metabolomics to map the glucuronidome in urine, serum, and colon/fecal samples from gnotobiotic and conventional mice. Our findings reveal microbiome-driven shifts in the glucuronidome, highlighting how differential GUS activity can influence host metabolite profiles. Reverse metabolomics of known glucuronidated chemicals and glucuronidation pattern filtering searches in public metabolomics datasets exposed the diversity of glucuronidated metabolites in human and mouse ecosystems. In summary, we present a new glucuronidation fingerprint resource that provides broader access to and analysis of the glucuronidome. By systematically capturing glucuronidation patterns, this resource enhances unknown metabolite annotation efforts and provides new insights into the dynamic relationship between the host and bacterial biotransformation activities.
Conflict of interest statement
Disclosures: PCD is an advisor and holds equity in Cybele, Sirenas, and BileOmix, and he is a scientific co-founder, advisor, holds equity and/or receives income from Ometa, Enveda, and Arome with prior approval by UC San Diego. PCD consulted for DSM Animal Health in 2023. MRR is a founder of Symberix, Inc., and has received funding from Merck and Lilly.
Figures






References
-
- Acker D. (2024). gg3D: 3D perspective plots for ggplot2. https://github.com/AckerDWM/gg3D
-
- Aggarwal H., Pathak P., Singh V., Kumar Y., Shankar M., Das B., Jagavelu K., & Dikshit M. (2021). Vancomycin-induced modulation of gram-positive gut bacteria and metabolites remediates insulin resistance in iNOS knockout mice. Frontiers in Cellular and Infection Microbiology, 11, 795333. 10.3389/fcimb.2021.795333 - DOI - PMC - PubMed
-
- Aron A. T., Gentry E. C., McPhail K. L., Nothias L.-F., Nothias-Esposito M., Bouslimani A., Petras D., Gauglitz J. M., Sikora N., Vargas F., van der Hooft J. J. J., Ernst M., Kang K. B., Aceves C. M., Caraballo-Rodríguez A. M., Koester I., Weldon K. C., Bertrand S., Roullier C., … Dorrestein P. C. (2020). Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nature Protocols, 15(6), 1954–1991. 10.1038/s41596-020-0317-5 - DOI - PubMed
-
- Asano Y., Hiramoto T., Nishino R., Aiba Y., Kimura T., Yoshihara K., Koga Y., & Sudo N. (2012). Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. American Journal of Physiology-Gastrointestinal and Liver Physiology, 303(11), G1288–G1295. 10.1152/ajpgi.00341.2012 - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

