A Method to Visualize Cell Proliferation of Arabidopsis thaliana: A Case Study of the Root Apical Meristem
- PMID: 40297840
- PMCID: PMC12037192
- DOI: 10.1002/pld3.70060
A Method to Visualize Cell Proliferation of Arabidopsis thaliana: A Case Study of the Root Apical Meristem
Abstract
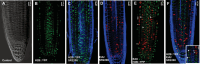
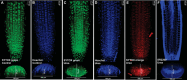
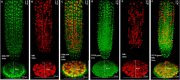
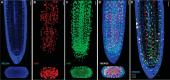
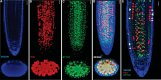
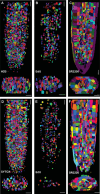
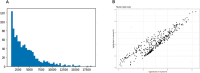
Plant growth and development rely on a delicate balance between cell proliferation and cell differentiation. The root apical meristem (RAM) of Arabidopsis thaliana is an excellent model to study the cell cycle due to the coordinated relationship between nucleus shape and cell size at each stage, allowing for precise estimation of the cell cycle duration. In this study, we present a method for high-resolution visualization of RAM cells. This is the first protocol that allows for simultaneous high-resolution imaging of cellular and nuclear stains, being compatible with DNA replication markers such as EdU, including fluorescent proteins (H2B::YFP), SYTOX DNA stains, and the cell wall stain SR2200. This protocol includes a clarification procedure that enables the acquisition of high-resolution 3D images, suitable for detailed subsequent analysis.
Keywords: 5‐ethynyl‐2‐deoxyuridine (EdU); cell and nuclear markers; cell cycle; root apical meristem.
© 2025 The Author(s). Plant Direct published by American Society of Plant Biologists and the Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Alberts, B. , Johnson A., Lewis J., et al. 2017. Molecular Biology of the Cell. 7th ed. New York: Garland Science.
-
- Bucevičius, J. , Lukinavičius G., and Gerasimaitė R.. 2018. “The Use of Hoechst Dyes for DNA Staining and Beyond.” Chem 6: 18.
LinkOut - more resources
Full Text Sources

