Discovery and biological evaluation of a novel and highly potent JAK2 inhibitor for the treatment of triple negative breast cancer
- PMID: 40298145
- PMCID: PMC12042240
- DOI: 10.1080/14756366.2025.2488127
Discovery and biological evaluation of a novel and highly potent JAK2 inhibitor for the treatment of triple negative breast cancer
Abstract
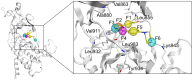
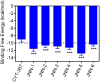
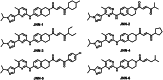
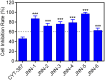
Janus kinase 2 (JAK2) is considered an attractive target for the treatment of triple-negative breast cancer (TNBC). Herein, we discovered six JAK2 inhibitors using structure-based virtual screening and molecular docking. Among them, JNN-5 was the best compound. It indicated strong inhibitory effects on JAK2 in the nanomolar range (IC50 = 0.41 ± 0.03 nM), and high selectivity for JAK2 over JAK1 and JAK3 (selectivity index (SI) > 73.17). Moreover, molecular dynamics (MD) simulation exhibited that JNN-5 bound with high stability to JAK2 JH1. Cellular assays revealed that JNN-5 displayed strong antiproliferative activities in the TNBC cell lines (MDA-MB-468, MDA-MB-213, HCC70, MDA-MB-157). JNN-5 significantly reduced the migration of HUVECs with the dose-dependence. JNN-5 had a significant inhibitory effect on multidrug-resistant MDA-MB-231/ADR (IC50 = 0.37 ± 0.02 μM). These data demonstrate that JNN-5 may be a highly effective and selective antitumor compound for the treatment of TNBC.
Keywords: JAK2; Triple negative breast cancer (TNBC); biological evaluation; inhibitor; structure-based virtual screening.
Conflict of interest statement
The authors report no conflicts of interest.
Figures


Similar articles
-
Chalcone-9: a novel inhibitor of the JAK-STAT pathway with potent anti-cancer effects in triple-negative breast cancer cells.Pharmacol Rep. 2025 Jun;77(3):761-774. doi: 10.1007/s43440-025-00721-w. Epub 2025 Apr 9. Pharmacol Rep. 2025. PMID: 40199813 Free PMC article.
-
Discovery of a novel small-molecule inhibitor of Fam20C that induces apoptosis and inhibits migration in triple negative breast cancer.Eur J Med Chem. 2021 Jan 15;210:113088. doi: 10.1016/j.ejmech.2020.113088. Epub 2020 Dec 7. Eur J Med Chem. 2021. PMID: 33316691
-
Discovery and characterization of novel FGFR1 inhibitors in triple-negative breast cancer via hybrid virtual screening and molecular dynamics simulations.Bioorg Chem. 2024 Sep;150:107553. doi: 10.1016/j.bioorg.2024.107553. Epub 2024 Jun 10. Bioorg Chem. 2024. PMID: 38901279
-
Inhibitors of JAK2 and JAK3: an update on the patent literature 2010 - 2012.Expert Opin Ther Pat. 2013 Apr;23(4):449-501. doi: 10.1517/13543776.2013.765862. Epub 2013 Feb 1. Expert Opin Ther Pat. 2013. PMID: 23367873 Review.
-
Novel benzothiazole-based dual VEGFR-2/EGFR inhibitors targeting breast and liver cancers: Synthesis, cytotoxic activity, QSAR and molecular docking studies.Bioorg Med Chem Lett. 2022 Feb 15;58:128529. doi: 10.1016/j.bmcl.2022.128529. Epub 2022 Jan 7. Bioorg Med Chem Lett. 2022. PMID: 35007724 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
