Biogeography and ecological functions of underestimated CPR and DPANN in acid mine drainage sediments
- PMID: 40298441
- PMCID: PMC12153355
- DOI: 10.1128/mbio.00705-25
Biogeography and ecological functions of underestimated CPR and DPANN in acid mine drainage sediments
Abstract
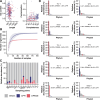
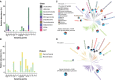
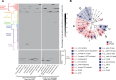
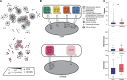
Recent genomic surveys have uncovered candidate phyla radiation (CPR) bacteria and DPANN archaea as major microbial dark matter lineages in various anoxic habitats. Despite their extraordinary diversity, the biogeographic patterns and ecological implications of these ultra-small and putatively symbiotic microorganisms have remained elusive. Here, we performed metagenomic sequencing on 90 geochemically diverse acid mine drainage sediments sampled across southeast China and recovered 282 CPR and 189 DPANN nonredundant metagenome-assembled genomes, which collectively account for up to 28.6% and 31.2% of the indigenous prokaryotic communities, respectively. We found that, remarkably, geographic distance represents the primary factor driving the large-scale ecological distribution of both CPR and DPANN organisms, followed by pH and Fe. Although both groups might be capable of iron reduction through a flavin-based extracellular electron transfer mechanism, significant differences are found in their metabolic capabilities (with complex carbon degradation and chitin degradation being more prevalent in CPR whereas fermentation and acetate production being enriched in DPANN), indicating potential niche differentiation. Predicted hosts are mainly Acidobacteriota, Bacteroidota, and Proteobacteria for CPR and Thermoplasmatota for DPANN, and extensive, unbalanced metabolic exchanges between these symbionts and putative hosts are displayed. Together, our results provide initial insights into the complex interplays between the two lineages and their physicochemical environments and host populations at a large geographic scale.IMPORTANCECandidate phyla radiation (CPR) bacteria and DPANN archaea constitute a significant fraction of Earth's prokaryotic diversity. Despite their ubiquity and abundance, especially in anoxic habitats, we know little about the community patterns and ecological drivers of these ultra-small, putatively episymbiotic microorganisms across geographic ranges. This study is facilitated by a large collection of CPR and DPANN metagenome-assembled genomes recovered from the metagenomes of 90 sediments sampled from geochemically diverse acid mine drainage (AMD) environments across southeast China. Our comprehensive analyses have allowed first insights into the biogeographic patterns and functional differentiation of these major enigmatic prokaryotic groups in the AMD model system.
Keywords: DPANN archaea; biogeography; host-cell interactions; microbial ecology; ultramicrobacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Genome-resolved metagenomics reveals site-specific diversity of episymbiotic CPR bacteria and DPANN archaea in groundwater ecosystems.Nat Microbiol. 2021 Mar;6(3):354-365. doi: 10.1038/s41564-020-00840-5. Epub 2021 Jan 25. Nat Microbiol. 2021. PMID: 33495623 Free PMC article.
-
Uncultivated DPANN archaea are ubiquitous inhabitants of global oxygen-deficient zones with diverse metabolic potential.mBio. 2024 Mar 13;15(3):e0291823. doi: 10.1128/mbio.02918-23. Epub 2024 Feb 21. mBio. 2024. PMID: 38380943 Free PMC article.
-
CPR bacteria and DPANN archaea play pivotal roles in response of microbial community to antibiotic stress in groundwater.Water Res. 2024 Mar 1;251:121137. doi: 10.1016/j.watres.2024.121137. Epub 2024 Jan 13. Water Res. 2024. PMID: 38246077
-
Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life.Cell. 2018 Mar 8;172(6):1181-1197. doi: 10.1016/j.cell.2018.02.016. Cell. 2018. PMID: 29522741 Review.
-
Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations.Nat Rev Microbiol. 2018 Oct;16(10):629-645. doi: 10.1038/s41579-018-0076-2. Nat Rev Microbiol. 2018. PMID: 30181663 Review.
References
-
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352 - DOI - PubMed
-
- Castelle CJ, Wrighton KC, Thomas BC, Hug LA, Brown CT, Wilkins MJ, Frischkorn KR, Tringe SG, Singh A, Markillie LM, Taylor RC, Williams KH, Banfield JF. 2015. Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol 25:690–701. doi: 10.1016/j.cub.2015.01.014 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- no. 31870111/National Natural Science Foundation of China
- no. 41830318/National Natural Science Foundation of China
- no. 32300001/National Natural Science Foundation of China
- no. 2022A1515010625/Natural Science Foundation of Guangdong Province
- no. 2021A1515012468/Natural Science Foundation of Guangdong Province
LinkOut - more resources
Full Text Sources

