Targeted Deletion of Cxcl1 in MSCs Regulates Osteogenesis and Suppresses Bone-Metastatic Prostate Cancer
- PMID: 40298918
- PMCID: PMC12319404
- DOI: 10.1158/1541-7786.MCR-24-0672
Targeted Deletion of Cxcl1 in MSCs Regulates Osteogenesis and Suppresses Bone-Metastatic Prostate Cancer
Abstract
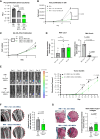
Bone metastasis continues to be the greatest challenge in treating patients with prostate cancer despite ongoing research. In bone, prostate cancer tumors hijack normal bone remodeling processes to drive cancer progression. However, it is unclear how these interactions drive bone-metastatic prostate cancer growth in the bone environment. To understand the mechanisms associated with bone-metastatic prostate cancer regulation of mesenchymal stem cells (MSC), we previously identified that bone-metastatic prostate cancer induces MSC expression of the pro-inflammatory chemokine CXCL8 and its mouse functional homologue Cxcl1. To date, there has been little to no information about the role of CXCL1/8 in MSC biology and its impact in the tumor-bone environment. Using genetic deletion of Cxcl1, we discovered a novel role for Cxcl1/8 in regulating MSC osteoblast differentiation, such that targeted deletion of Cxcl1 enhanced MSC osteoblastogenesis. Despite the osteogenic nature of prostate cancer, co-injection of Cxcl1 knockout (KO) MSCs with bone-metastatic prostate cancer in bone significantly suppressed tumor growth compared with co-injection with scrambled control (non-targeting) MSCs, even in the presence of three times more prostate cancer to MSCs. Furthermore, bulk RNA sequencing revealed immune response pathways, both in Cxcl1-KO MSCs and bone-metastatic prostate cancer tumors containing Cxcl1-KO MSCs. In support of this, Cxcl1-KO MSCs reduced immature neutrophils in the bone environment, while increasing monocytes. These findings demonstrate the importance of MSC-derived Cxcl1 in the bone microenvironment and highlight the importance of Cxcl1 in bone-metastatic prostate cancer progression.
Implications: MSC-derived Cxcl1 regulates prostate cancer progression in bone.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
S. Rajgopal reports grants from UNMC and other support from NCI during the conduct of the study. J.S. Frieling reports grants from H. Lee Moffitt Cancer Center & Research Institute during the conduct of the study. L.M. Cook reports grants from American Cancer Society and other support from NCI during the conduct of the study. No disclosures were reported by the other authors.
Figures




References
-
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin 2024;74:12–49. - PubMed
-
- Achard V, Putora PM, Omlin A, Zilli T, Fischer S. Metastatic prostate cancer: treatment options. Oncology 2022;100:48–59. - PubMed
-
- Sturge J, Caley MP, Waxman J. Bone metastasis in prostate cancer: emerging therapeutic strategies. Nat Rev Clin Oncol 2011;8:357–68. - PubMed
MeSH terms
Substances
Grants and funding
- RSG-19-127-01-CSM/American Cancer Society (ACS)
- R01 CA274605/CA/NCI NIH HHS/United States
- I01 BX005886/BX/BLRD VA/United States
- IK6 BX005962/BX/BLRD VA/United States
- 1R01CA274605-01/National Cancer Institute (NCI)
- P30 CA036727/CA/NCI NIH HHS/United States
- P50 AA030407/AA/NIAAA NIH HHS/United States
- ACORN P50 AA030407/National Institutes of Health (NIH)
- T32 CA009476/CA/NCI NIH HHS/United States
- T32CA009476/National Cancer Institute (NCI)
- IK6 BX003781/BX/BLRD VA/United States
- P20 GM103427/GM/NIGMS NIH HHS/United States
- ZIA BC012170/ImNIH/Intramural NIH HHS/United States
- IS1 BX004790/BX/BLRD VA/United States
- Z99 CA999999/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

