Integration of Transcriptomic and Single-Cell Data to Uncover Senescence- and Ferroptosis-Associated Biomarkers in Sepsis
- PMID: 40299574
- PMCID: PMC12025025
- DOI: 10.3390/biomedicines13040942
Integration of Transcriptomic and Single-Cell Data to Uncover Senescence- and Ferroptosis-Associated Biomarkers in Sepsis
Abstract
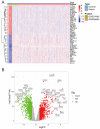
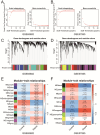
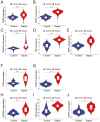
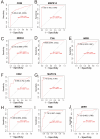
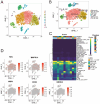
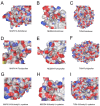
Background: Sepsis is a life-threatening condition characterized by organ dysfunction due to an imbalanced immune response to infection, with high mortality. Ferroptosis, an iron-dependent cell death process, and cellular senescence, which exacerbates inflammation, have recently been implicated in sepsis pathophysiology. Methods: Weighted gene co-expression network analysis (WGCNA) was used to identify ferroptosis- and senescence-related gene modules in sepsis. Differentially expressed genes (DEGs) were analyzed using public datasets (GSE57065, GSE65682, and GSE26378). Receiver operating characteristic (ROC) analysis was performed to evaluate their diagnostic potential, while single-cell RNA sequencing (scRNA-seq) was used to assess their immune-cell-specific expression. Molecular docking was conducted to predict drug interactions with key proteins. Results: Five key genes (CD82, MAPK14, NEDD4, TXN, and WIPI1) were significantly upregulated in sepsis patients and highly correlated with immune cell infiltration. MAPK14 and TXN exhibited strong diagnostic potential (AUC = 0.983, 0.978). Molecular docking suggested potential therapeutic interactions with diclofenac, flurbiprofen, and N-acetyl-L-cysteine. Conclusions: This study highlights ferroptosis and senescence as critical mechanisms in sepsis and identifies promising biomarkers for diagnosis and targeted therapy. Future studies should focus on clinical validation and precision medicine applications.
Keywords: ferroptosis; senescence; sepsis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.-D., Coopersmith C.M., et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. - DOI - PMC - PubMed
-
- Rudd K.E., Johnson S.C., Agesa K.M., Shackelford K.A., Tsoi D., Kievlan D.R., Colombara D.V., Ikuta K.S., Kissoon N., Finfer S., et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet. 2020;395:200–211. doi: 10.1016/S0140-6736(19)32989-7. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

