Rice stripe mosaic virus M protein antagonizes G-protein-induced antiviral autophagy in insect vectors
- PMID: 40300033
- PMCID: PMC12040238
- DOI: 10.1371/journal.ppat.1013070
Rice stripe mosaic virus M protein antagonizes G-protein-induced antiviral autophagy in insect vectors
Abstract
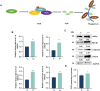
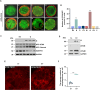
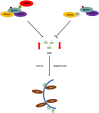
In the field, 80% of plant viruses are transmitted by insect vectors. When ingested by a sap-sucking insect such as Recilia dorsalis, persistently transmitted viruses such as rice stripe mosaic virus (RSMV) infect the gut epithelium and eventually pass to the salivary glands where they will be transmitted to the next rice (Oryza sativa) plant. To efficiently exploit insect vectors for transmission, plant viruses must overcome various immune mechanisms within the vectors, including autophagy. However, understanding how plant viruses overcome insect autophagic defenses remains limited. In this study, we provide evidence that infection with RSMV triggers an autophagic antiviral response in leafhopper cells. In this response, the G protein of RSMV binds to a leafhopper AMP-activated protein kinase (AMPK), leading to enhanced phosphorylation of Beclin-1 (BECN1), thereby inducing autophagy. Knockdown of AMPK and genes encoding members of the phosphoinositide 3-kinase (PI3K) complex composed of the autophagy-related protein 14 (ATG14), BECN1, and vacuolar protein sorting 34 (VPS34) facilitated viral infection in leafhoppers. To suppress leafhopper-induced autophagy, RSMV M protein specifically interacts with ATG14, resulting in the disintegration of PI3K complexes. This leads to reduced phosphatidylinositol-3-phosphate content and thus inhibits the G-protein- induced autophagy. Our study sheds light on the mechanism by which this rice virus evades insect autophagy antiviral defenses.
Copyright: © 2025 Zhang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Zhang Y, Ju F. Fighting caterpillar pests and managing agricultural insecticide resistance with Lepidoptera-associated Enterococcus casseliflavus. TIL. 2023;1(3):100042. doi: 10.59717/j.xinn-life.2023.100042 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

