Efficacy of ATR Kinase Inhibitor Elimusertib Monotherapy or Combination in Tumors with DNA Damage Response Pathway and Other Genomic Alterations
- PMID: 40300249
- PMCID: PMC12402799
- DOI: 10.1158/1535-7163.MCT-24-0884
Efficacy of ATR Kinase Inhibitor Elimusertib Monotherapy or Combination in Tumors with DNA Damage Response Pathway and Other Genomic Alterations
Abstract
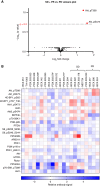
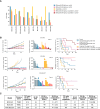
The ataxia telangiectasia and RAD3-related (ATR) kinase functions with ataxia telangiectasia-mutated (ATM) kinase as a modulator of DNA damage response (DDR). We assessed the antitumor effects of the ATR inhibitor elimusertib (BAY-1895344) in patient-derived xenograft (PDX) models with DDR alterations. Antitumor activity was assessed by change in tumor volume (TV) from baseline. Responses were categorized as follows: partial response (PR), ≥30% decrease in TV; ≥20% increase in TV, progressive disease; and non-PR/progressive disease, stable disease (SD). Event-free survival was defined as time for tumor doubling (EFS-2). Of 21 PDX models tested, 11 had significant prolongation of EFS-2 with elimusertib monotherapy. Four models had a PR and four had SD. PR/SD was observed in two of five models with ATM loss on IHC and in models with a variety of alterations in DDR genes, including BRCA1/2 and ATM. Elimusertib prolonged EFS-2 in three of five models with known PARP inhibitor resistance. Pharmacodynamic studies conducted in four PDX models showed an increase in DNA damage markers. PI3K/mTOR pathway signaling increased in two of four models. The combination of the PI3K inhibitor copanlisib with elimusertib enhanced EFS-2 compared with monotherapy in three of 11 models tested. The combination of elimusertib with the PARP inhibitor niraparib enhanced antitumor activity compared with single agents in PARP-resistant PDX models. Our study shows that ATR inhibition has antitumor activity, including in models with both intrinsic and acquired PARP inhibitor resistance. Further work is needed to better refine patient selection for ATR-based therapies.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
K.W. Evans reports grants from Bayer during the conduct of the study. A.M. Wengner reports employment with Bayer AG at the time of data generation. T.A. Yap reports other support from University of Texas MD Anderson Cancer Center (VP, Head of Clinical Development in the Therapeutics Discovery Division, which has a commercial interest in drug discovery and development), personal fees from AbbVie, Acrivon, Adagene, Aeneid Therapeutics, Almac, Alterome Therapeutics Inc., Audro, Amgen Inc., Amphista, Astex, Atavistik, Athena, Atrin, Avenzo, Avoro, Axiom, Baptist Health Systems, Bicycle, BioCity Pharma, Bluestar Bio, Boxer, Bristol Myers Squibb, C4 Therapeutics, Calithera, Cancer Research Horizons, Cancer Research UK, Carrick Therapeuitcs, Circle Pharma, Clasp, Cybrexa, Daiichi Sankyo, DAiNA, Dark Blue Therapeutics, Dawn Manco, Debiopharm, Diffusion, Duke Street Bio, EcoR1 Capital, Eikon, Ellipses Pharma, Entos, Flagship Pioneering, Forbion, FoRx Therapeutics, Genesis Therapeutics, Genmab, Glenmark, GLG, Globe Life Sciences, Grey Wolf Therapeutics, GSK, Guardant, Guidepoint, Idience, Ignyta, I-Mab, Impact Therapeutics, Institut Gustave Roussy, Intellisphere, Janssen, Jazz Pharma, Joint Scientific Committee for Phase I Trials in Hong Kong, Kyn, Kyowa Kirin, Lumanity, MEI pharma, Mereo, Merit, Monte Rosa Therapeutics, Natera, Nested Therapeutics, Nexus Pharmaceuticals, Nimbus, Novocure, Odyssey Therapeutics, OHSU, OncoSec, Ono Pharma, Onxeo, PanAngium Therapeutics, Pegascy, PER, Pfizer, Piper-Sandler, Pliant Therapeutics, Plexium Inc., Prelude Therapeutics, Prolynx, Protai Bio, PSIM, Radiopharma Theranostics, Repare, resTORbio, Roche, Ryvu Therapeutics, SAKK, Sanofi, Schrodinger, Servier, Stablix, Synnovation, Synthis Therapeutics, Tango, TCG Crossover, TD2, Techspert.io, Terremoto Biosciences, Tessellate Bio, Theragnostics, Terns Pharmaceuticals, Thryv Therapeutics, Tolremo, Tome Biosciences, Trevarx Biomedical, Varian, Veeva, Versant, Vibliome Therapeutics, Vivace, Voronoi Inc., Xinthera, and Zai Labs, grants and personal fees from Artios, AstraZeneca, Bayer, BeiGene, Blueprint, BridGene Biosciences, Clovis, 858 Therapeutics, EMD Serono, F-Star, Ideaya Biosciences, ImmuneSensor, and Merck, and grants from Accent, Aprea Therapeutics, BioNTech, Bristol Myers Squibb, Boundless Bio, Circle Pharma, Constellation, CPRIT, Cyteir, Department of Defense, Eisbach Bio, Eli Lilly and Company, Exelixis, Forbius, Gilead Sciences, GlaxoSmithKline, Genetech, Golfers Against Cancer, Haihe, Insilico Medicine, Ionis, Ipsen, Jounce, Karyopharm, KSQ, Kyowa, Loxo Oncology, Mirati, Novartis, NIH/NCI, Pfizer, Pliant, Prelude, Ribon Therapeutics, Regeneron, Repare, Roche, Rubius, Sanofi, Scholar Rock, Seattle Genetics, Springworks, Synnovation, Tango, Tesaro, V Foundation, Zenith, Vivace, and Zentalis outside the submitted work. F. Meric-Bernstam reports grants and personal fees from AstraZeneca, Daiichi Sankyo, Debiopharm, eFFECTOR Therapeutics, Zymeworks, and Guardant Health, personal fees from Becton Dickinson, Calibr (a division of Scripps Research), Dava Oncology, EcoR1 Capital, Elevation Oncology, Exelixis, GT Aperion, Incyte, Jazz Pharmaceuticals, LegoChem Biosciences, Lengo Therapeutics, Menarini Group, Molecular Templates, Protai Bio, Ribometrix, Tallac Therapeutics, Tempus, Cybrexa, FogPharma, GO Therapeutics, Harbinger Health, Karyopharm Therapeutics, Kivu Biosciences, LOXO Oncology, Mersana Therapeutics, OnCusp Therapeutics, Sanofi, Seagan, Theratechnologies, and Zentalis Pharmaceuticals, grants from Jazz Pharmaceuticals, Aileron Therapeutics, Bayer Healthcare, Calithera Biosciences, Inc., Curis, Inc., CytomX Therapeutics, Genentech, Klus Pharma, Takeda Pharmaceutical, Novartis, Puma Biotechnology, Inc., and Taiho Pharmaceutical, personal fees and other support from Dava Oncology, and other support from European Organisation for Research and Treatment of Cancer, European Society for Medical Oncology, and Cholangiocarcinoma Foundation outside the submitted work. No disclosures were reported by the other authors.
Figures




Similar articles
-
Factors Influencing the Central Nervous System (CNS) Distribution of the Ataxia Telangiectasia Mutated and Rad3-Related Inhibitor Elimusertib (BAY1895344): Implications for the Treatment of CNS Tumors.J Pharmacol Exp Ther. 2024 Oct 18;391(2):346-360. doi: 10.1124/jpet.123.002002. J Pharmacol Exp Ther. 2024. PMID: 39284626
-
ATR inhibition increases reliance on PARP-mediated DNA repair revealing an improved therapeutic strategy for cervical cancer.Gynecol Oncol. 2024 Dec;191:182-193. doi: 10.1016/j.ygyno.2024.10.009. Epub 2024 Oct 19. Gynecol Oncol. 2024. PMID: 39427557
-
Phase I trial of ATR inhibitor elimusertib with FOLFIRI in advanced or metastatic gastrointestinal malignancies (ETCTN 10406).Cancer Chemother Pharmacol. 2025 Jan 22;95(1):27. doi: 10.1007/s00280-024-04745-6. Cancer Chemother Pharmacol. 2025. PMID: 39841295 Clinical Trial.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Rho kinase inhibitor for primary open-angle glaucoma and ocular hypertension.Cochrane Database Syst Rev. 2022 Jun 10;6(6):CD013817. doi: 10.1002/14651858.CD013817.pub2. Cochrane Database Syst Rev. 2022. PMID: 35686679 Free PMC article.
References
-
- Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell 2017;66:801–17. - PubMed
-
- Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther 2015;149:124–38. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. - PubMed
-
- Kwok M, Davies N, Agathanggelou A, Smith E, Oldreive C, Petermann E, et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood 2016;127:582–95. - PubMed
-
- Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 2011;7:428–30. - PubMed
MeSH terms
Substances
Grants and funding
- U54 CA224065/CA/NCI NIH HHS/United States
- U45 #CA224065/PDX Development and Trial Center
- UL1 TR003167/TR/NCATS NIH HHS/United States
- Barr Foundation (BF)
- Nellie B. Connally Breast Cancer Research Endowment
- 1UL1TR003167/National Center for Advancing Translational Sciences (NCATS)
- T32 CA009599/CA/NCI NIH HHS/United States
- CA009599/National Institutes of Health (NIH)
- P30 CA016672/CA/NCI NIH HHS/United States
- Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy (Khalifa Institute)
- MD Anderson Cancer Moonshot
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

