Targeting of the Hybrid Bamboo BDDnaJ by Pathogen Effector ApcE12 Regulates the Unfolded Protein Response
- PMID: 40300854
- PMCID: PMC12040440
- DOI: 10.1111/mpp.70089
Targeting of the Hybrid Bamboo BDDnaJ by Pathogen Effector ApcE12 Regulates the Unfolded Protein Response
Abstract
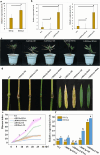
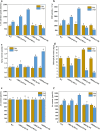
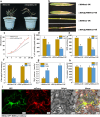
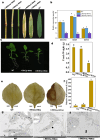
The shoot blight disease of Bambusa pervariabilis × Dendrocalamopsis grandis, caused by Arthrinium phaeospermum, threatens bamboo's ecological and economic value. This study explores the pathogenic effector ApcE12's role in modulating plant immunity through interactions with the host proteins BDClp and BDDnaJ. ApcE12 directly interacts with BDDnaJ, a vital regulator of the unfolded protein response (UPR), as validated through yeast two-hybrid, bimolecular fluorescence complementation, and GST pull-down assays. Functional analyses demonstrated that silencing BDDnaJ reduces UPR, activating programmed cell death (PCD) and blocking further pathogen infection to enhance plant resistance. BDDnaJ was found to regulate BDBiP protein stability by interacting with BDBiP, and it is this mechanism by which the pathogenic effector ApcE12 regulates BDDnaJ expression, enhances UPR signalling, and inhibits PCD, thereby promoting infection. These findings deepen our understanding of how fungal effectors manipulate UPR and PCD to overcome plant defences, providing novel insights for developing resistance strategies in bamboo species.
Keywords: Arthrinium phaeospermum; Bambusa pervariabilis × Dendrocalamopsis grandis; programmed cell death; protein–protein interactions; unfolded protein response.
© 2025 The Author(s). Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Functional Identification of Arthrinium phaeospermum Effectors Related to Bambusa pervariabilis × Dendrocalamopsis grandis Shoot Blight.Biomolecules. 2022 Sep 8;12(9):1264. doi: 10.3390/biom12091264. Biomolecules. 2022. PMID: 36139102 Free PMC article.
-
Verification of the Interaction Target Protein of the Effector ApCE22 of Arthrinium phaeospermum in Bambusa pervariabilis × Dendrocalamopsis grandis.Biomolecules. 2023 Mar 25;13(4):590. doi: 10.3390/biom13040590. Biomolecules. 2023. PMID: 37189340 Free PMC article.
-
Identification of the interacting proteins of Bambusa pervariabilis × Dendrocalamopsis grandis in response to the transcription factor ApCtf1β in Arthrinium phaeospermum.Front Plant Sci. 2022 Sep 15;13:991077. doi: 10.3389/fpls.2022.991077. eCollection 2022. Front Plant Sci. 2022. PMID: 36186076 Free PMC article.
-
UPR signaling at the nexus of plant viral, bacterial, and fungal defenses.Curr Opin Virol. 2021 Apr;47:9-17. doi: 10.1016/j.coviro.2020.11.001. Epub 2020 Dec 22. Curr Opin Virol. 2021. PMID: 33360330 Review.
-
The potential of effector-target genes in breeding for plant innate immunity.Microb Biotechnol. 2013 May;6(3):223-9. doi: 10.1111/1751-7915.12023. Epub 2012 Dec 27. Microb Biotechnol. 2013. PMID: 23279965 Free PMC article. Review.
References
-
- Ai, G. , Zhu H., Fu X., et al. 2021. “ Phytophthora Infection Signals‐Induced Translocation of NAC089 Is Required for Endoplasmic Reticulum Stress Response‐Mediated Plant Immunity.” Plant Journal 108: 67–80. - PubMed
-
- Boevink, P. C. , McLellan H., Gilroy E. M., et al. 2016. “Oomycetes Seek Help From the Plant: Phytophthora infestans Effectors Target Host Susceptibility Factors.” Molecular Plant 9: 636–638. - PubMed
MeSH terms
Substances
Grants and funding
- 32171795/National Natural Science Foundation of China
- 2023NSFSC1936/Sichuan Natural Science Foundation for Distinguished Young Scholar
- The Young Talents of Sichuan Tianfu Qingcheng Program
- 2025290/The Innovation Training Program for College Students
- 20251602/The Research Interest Training Program for College Students
LinkOut - more resources
Full Text Sources
Research Materials

