Immunopeptidomics identified antigens for mRNA-lipid nanoparticle vaccines with alpha-galactosylceramide in multiple myeloma therapy
- PMID: 40300855
- PMCID: PMC12049997
- DOI: 10.1136/jitc-2024-010673
Immunopeptidomics identified antigens for mRNA-lipid nanoparticle vaccines with alpha-galactosylceramide in multiple myeloma therapy
Abstract
Background: Invariant natural killer T (iNKT) cells and CD8+ T cells are key in the immune response against multiple myeloma (MM), a largely incurable blood cancer. Immunization is a promising strategy to activate these T cell populations. To our knowledge, immunization with messenger RNA (mRNA) and the iNKT agonist, α-galactosylceramide (αGC), has not been studied in MM, as knowledge on clinically relevant antigens in preclinical MM models is lacking.
Methods: Microarray data and immunopeptidomics (imPep) were used to identify candidate antigens for immunization in 5TMM models. Galsomes, lipid nanoparticles containing antigen mRNA and αGC were used to immunize 5T33MM-bearing mice. This treatment was combined with a CD40 agonist. Tumor burden and activation of iNKT cells and CD8+ T cells were studied using M-protein electrophoresis, flow cytometry and ELISA.
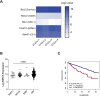
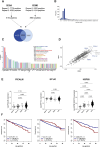
Results: RNA transcripts revealed survivin as a candidate antigen. Prime-boost Galsomes therapy targeting survivin significantly reduced M-protein levels despite low survivin-specific T cell responses. Further analysis showed potential T cell fratricide. ImPep revealed HSP60, Idiotype, PICALM and EF1A1 as candidate antigens. Prime-boost therapy with Galsomes targeting these antigens reduced MM growth significantly when combined with a CD40 agonist, coinciding with significantly improved antigen presentation, costimulation and cytotoxicity of iNKT cells and CD8+ T cells.
Conclusion: These findings highlight the potential of Galsomes, an mRNA vaccine designed to activate CD8+ T cells and iNKT cells, for MM therapy, and emphasize the importance of combinatorial approaches, addressing immune anergy for effective MM immunotherapies.
Keywords: Immunotherapy; Major histocompatibility complex - MHC; Multiple Myeloma; Nanoparticle; Vaccine.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: The authors declare the following competing interest: therapeutic nanoparticles and methods of use thereof is the topic of a patent (WO2020058239A1) on which IL an RV are inventors. None of the authors receive any support or remuneration related to this platform.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
