The aldehyde dehydrogenase superfamilies: correlations and deviations in structure and function
- PMID: 40301148
- PMCID: PMC12041015
- DOI: 10.1007/s00253-025-13467-5
The aldehyde dehydrogenase superfamilies: correlations and deviations in structure and function
Abstract
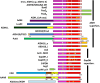
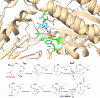
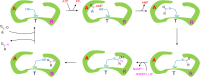
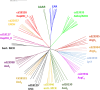
Aldehyde dehydrogenases participate in many biochemical pathways, either by degrading organic substrates via organic acids or by producing reactive aldehyde intermediates in many biosynthetic pathways, and are becoming increasingly important for constructing synthetic metabolic pathways. Although they consist of simple and highly conserved basic structural motifs, they exhibit a surprising variability in the reactions catalyzed. We attempt here to give an overview of the known enzymes of two superfamilies comprising the known aldehyde dehydrogenases, focusing on their structural similarities and the residues involved in the catalytic reactions. The analysis reveals that the enzymes of the two superfamilies share many common traits and probably have a common evolutionary origin. While all enzymes catalyzing irreversible aldehyde oxidation to acids exhibit a universally conserved reaction mechanism with shared catalytic active-site residues, the enzymes capable of reducing activated acids to aldehydes deviate from this mechanism, displaying different active-site modifications required to allow these reactions which apparently evolved independently in different enzyme subfamilies. KEY POINTS: • The two aldehyde dehydrogenase superfamilies share significant similarities. • Catalytic amino acids of irreversibly acting AlDH are universally conserved. • Reductive or reversible reactions are enabled by water exclusion via the loss of conserved residues.
Keywords: Aldehyde dehydrogenase; Catalytic mechanism; Conserved domains; Glyceraldehyde-3-phosphate; Structure–function relations.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: This article does not contain any studies with human participants or animals performed by the authors. Conflict of interest: The authors declare no competing interests.
Figures





References
-
- Arbore R, Barbosa S, Brejcha J, Ogawa Y, Liu Y, Nicolaï MPJ, Pereira P, Sabatino SJ, Cloutier A, Poon ESK, Marques CI, Andrade P, Debruyn G, Afonso S, Afonso R, Roy SG, Abdu U, Lopes RJ, Mojzeš P, Marsˇík P, Sin SYW, White MA, Araújo PM, Corbo JC, Carneiro M (2024) A molecular mechanism for bright color variation in parrots. Science 386:eadp7710. 10.1126/science.adp7710 - DOI - PMC - PubMed
-
- Baré G, Swiatkowski T, Moukil A, Gerday C, Thonart P (2002) Purification and characterization of a microbial dehydrogenase: a vanillin:NAD(P)+ oxidoreductase. Appl Biochem Biotechnol - Part A Enzym Eng Biotechnol 98–100:415–428. 10.1385/ABAB:98-100:1-9:415 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

