AI-driven discovery of synergistic drug combinations against pancreatic cancer
- PMID: 40301300
- PMCID: PMC12041571
- DOI: 10.1038/s41467-025-56818-6
AI-driven discovery of synergistic drug combinations against pancreatic cancer
Abstract
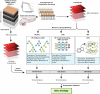
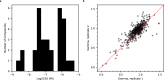
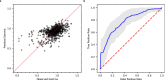
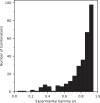
Pancreatic cancer treatment often relies on multi-drug regimens, but optimal combinations remain elusive. This study evaluates predictive approaches to identify synergistic drug combinations using a dataset from the National Center for Advancing Translational Sciences (NCATS). Screening 496 combinations of 32 anticancer compounds against the PANC-1 cells experimentally determined the degree of synergism and antagonism. Three research groups (NCATS, University of North Carolina, and Massachusetts Institute of Technology) leverage these data to apply machine learning (ML) approaches, predicting synergy across 1.6 million combinations. Of the 88 tested, 51 show synergy, with graph convolutional networks achieving the best hit rate and random forest the highest precision. Beyond highlighting the potential of ML, this work delivers 307 experimentally validated synergistic combinations, demonstrating its practical impact in treating pancreatic cancer.
© 2025. Massachusetts Institute of Technology and UNC Eshelman School of Pharmacy. Parts of this work were authored by US Federal Government authors and are not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: A.T. and E.N.M. are co-founders of Predictive, LLC, which develops novel alternative methodologies and software for toxicity prediction. The remaining authors declare no competing interests.
Figures


References
-
- Wang, C. et al. PANC-1 Pancreatic Cancer Cell Growth Inhibited by Cucurmosin Alone and in Combination With an Epidermal Growth Factor Receptor–Targeted Drug. Pancreas43, 291–297 (2014). - PubMed
-
- Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2015. Ca. Cancer J. Clin.65, 5–29 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

