Multipronged SMAD pathway targeting by lipophilic poly(β-amino ester) miR-590-3p nanomiRs inhibits mesenchymal glioblastoma growth and prolongs survival
- PMID: 40301302
- PMCID: PMC12041600
- DOI: 10.1038/s41392-025-02223-w
Multipronged SMAD pathway targeting by lipophilic poly(β-amino ester) miR-590-3p nanomiRs inhibits mesenchymal glioblastoma growth and prolongs survival
Abstract
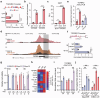
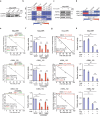
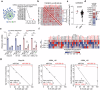
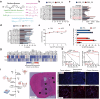
Despite aggressive therapy, glioblastoma (GBM) recurs in almost all patients and treatment options are very limited. Despite our growing understanding of how cellular transitions associate with relapse in GBM, critical gaps remain in our ability to block these molecular changes and treat recurrent disease. In this study we combine computational biology, forward-thinking understanding of miRNA biology and cutting-edge nucleic acid delivery vehicles to advance targeted therapeutics for GBM. Computational analysis of RNA sequencing from clinical GBM specimens identified TGFβ type II receptor (TGFBR2) as a key player in the mesenchymal transition associated with worse outcome in GBM. Mechanistically, we show that elevated levels of TGFBR2 is conducive to reduced temozolomide (TMZ) sensitivity. This effect is, at least partially, induced by stem-cell driving events coordinated by the reprogramming transcription factors Oct4 and Sox2 that lead to open chromatin states. We show that blocking TGFBR2 via molecular and pharmacological approaches decreases stem cell capacity and sensitivity of clinical recurrent GBM (rGBM) isolates to TMZ in vitro. Network analysis uncovered miR-590-3p as a tumor suppressor that simultaneously inhibits multiple oncogenic nodes downstream of TGFBR2. We also developed novel biodegradable lipophilic poly(β-amino ester) nanoparticles (LiPBAEs) for in vivo microRNA (miRNAs) delivery. Following direct intra-tumoral infusion, these nanomiRs efficiently distribute through the tumors. Importantly, miR-590-3p nanomiRs inhibited the growth and extended survival of mice bearing orthotopic human rGBM xenografts, with an apparent 30% cure rate. These results show that miRNA-based targeted therapeutics provide new opportunities to treat rGBM and bypass the resistance to standard of care therapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.G. and S.T. are managers and co-founders of OncoSwitch. J.G. is a board member, CSO, and co-founder of Cove Therapeutics and a manager, CTO, and co-founder of Dome Therapeutics. Patents related to the technology discussed in the manuscript and any potential conflicts of interest are managed by the Johns Hopkins University Committee on Outside Interests. There are no competing interests from the other authors.
Figures


References
-
- Birch, J. L., et al. Multifaceted transforming growth factor-beta (TGFbeta) signalling in glioblastoma. Cell Signal72, 109638 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- R01 NS096754/NS/NINDS NIH HHS/United States
- R37CA246699/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- 5P50HD103538-05/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- P30 CA006973/CA/NCI NIH HHS/United States
- R01NS096754/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- P30CA006973/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01CA228133/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 NS073611/NS/NINDS NIH HHS/United States
- S10 OD016374/OD/NIH HHS/United States
- F99CA284254/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P50 HD103538/HD/NICHD NIH HHS/United States
- R37 CA246699/CA/NCI NIH HHS/United States
- R01NS073611/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- S10 OD016374/CD/ODCDC CDC HHS/United States
- R01 NS120949/NS/NINDS NIH HHS/United States
- R01 CA228133/CA/NCI NIH HHS/United States
- P41EB028239/U.S. Department of Health & Human Services | NIH | National Institute of Biomedical Imaging and Bioengineering (NIBIB)
- F99 CA284254/CA/NCI NIH HHS/United States
- P41 EB028239/EB/NIBIB NIH HHS/United States
- R01NS120949/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
LinkOut - more resources
Full Text Sources
Medical

