Autophagy disruption and mitochondrial stress precede photoreceptor necroptosis in multiple mouse models of inherited retinal disorders
- PMID: 40301324
- PMCID: PMC12041483
- DOI: 10.1038/s41467-025-59165-8
Autophagy disruption and mitochondrial stress precede photoreceptor necroptosis in multiple mouse models of inherited retinal disorders
Abstract
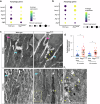
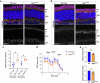
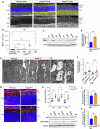
Inherited retinal diseases (IRDs) are a leading cause of blindness worldwide. One of the greatest barriers to developing treatments for IRDs is the heterogeneity of these disorders, with causative mutations identified in over 280 genes. It is therefore a priority to find therapies applicable to a broad range of genetic causes. To do so requires a greater understanding of the common or overlapping molecular pathways that lead to photoreceptor death in IRDs and the molecular processes through which they converge. Here, we characterise the contribution of different cell death mechanisms to photoreceptor degeneration and loss throughout disease progression in humanised mouse models of IRDs. Using single-cell transcriptomics, we identify common transcriptional signatures in degenerating photoreceptors. Further, we show that in genetically and functionally distinct IRD models, common early defects in autophagy and mitochondrial damage exist, triggering photoreceptor cell death by necroptosis in later disease stages. These results suggest that, regardless of the underlying genetic cause, these pathways likely contribute to cell death in IRDs. These insights provide potential therapeutic targets for novel, gene-agnostic treatments for IRDs applicable to the majority of patients.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Neogenin neutralization prevents photoreceptor loss in inherited retinal degeneration.J Clin Invest. 2020 Apr 1;130(4):2054-2068. doi: 10.1172/JCI125898. J Clin Invest. 2020. PMID: 32175920 Free PMC article.
-
cAMP and Photoreceptor Cell Death in Retinal Degeneration.Adv Exp Med Biol. 2019;1185:301-304. doi: 10.1007/978-3-030-27378-1_49. Adv Exp Med Biol. 2019. PMID: 31884628 Review.
-
Primary and Secondary Cone Cell Death Mechanisms in Inherited Retinal Diseases and Potential Treatment Options.Int J Mol Sci. 2022 Jan 10;23(2):726. doi: 10.3390/ijms23020726. Int J Mol Sci. 2022. PMID: 35054919 Free PMC article. Review.
-
Multiple, parallel cellular suicide mechanisms participate in photoreceptor cell death.Exp Eye Res. 2006 Aug;83(2):380-9. doi: 10.1016/j.exer.2006.01.014. Epub 2006 Apr 19. Exp Eye Res. 2006. PMID: 16626700
-
A Role for SARM1 in Photoreceptor Cell Death.Adv Exp Med Biol. 2025;1468:183-187. doi: 10.1007/978-3-031-76550-6_30. Adv Exp Med Biol. 2025. PMID: 39930193 Review.
References
-
- Wright, A. F., Chakarova, C. F., Abd El-Aziz, M. M. & Bhattacharya, S. S. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat. Rev. Genet.11, 273–284 (2010). - PubMed
-
- RetNet. Retinal Information Network. https://web.sph.uth.edu/RetNet/ (2024).
-
- Yanik, M. et al. vivo genome editing as a potential treatment strategy for inherited retinal dystrophies. Prog. Retin. Eye Res.56, 1–18 (2017). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

