Glycosylation of serine/threonine-rich intrinsically disordered regions of membrane-associated proteins in streptococci
- PMID: 40301326
- PMCID: PMC12041528
- DOI: 10.1038/s41467-025-58692-8
Glycosylation of serine/threonine-rich intrinsically disordered regions of membrane-associated proteins in streptococci
Abstract
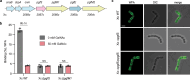
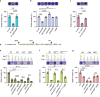
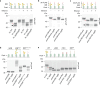
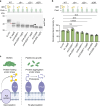
Proteins harboring intrinsically disordered regions (IDRs) lacking stable secondary or tertiary structures are abundant across the three domains of life. These regions have not been systematically studied in prokaryotes. Here, our genome-wide analysis identifies extracytoplasmic serine/threonine-rich IDRs in several biologically important membrane-associated proteins in streptococci. We demonstrate that these IDRs are glycosylated with glucose by glycosyltransferases GtrB and PgtC2 in Streptococcus pyogenes and Streptococcus pneumoniae, and with N-acetylgalactosamine by a Pgf-dependent mechanism in Streptococcus mutans. The absence of glycosylation leads to a defect in biofilm formation under ethanol-stressed conditions in S. mutans. We link this phenotype to the C-terminal IDR of the post-translocation chaperone PrsA. Our data reveal that O-linked glycosylation protects the IDR-containing proteins from proteolytic degradation and is critical for the biological function of PrsA in biofilm formation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Update of
-
Glycosylation of serine/threonine-rich intrinsically disordered regions of membrane-associated proteins in streptococci.bioRxiv [Preprint]. 2025 Mar 17:2024.05.05.592596. doi: 10.1101/2024.05.05.592596. bioRxiv. 2025. Update in: Nat Commun. 2025 Apr 29;16(1):4011. doi: 10.1038/s41467-025-58692-8. PMID: 38746434 Free PMC article. Updated. Preprint.
References
-
- Uversky, V. N. Functional unfoldomics: Roles of intrinsic disorder in protein (multi)functionality. Adv. Protein Chem. Struct. Biol.138, 179–210 (2024). - PubMed
-
- Ward, J. J., Sodhi, J. S., McGuffin, L. J., Buxton, B. F. & Jones, D. T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol.337, 635–645 (2004). - PubMed
MeSH terms
Substances
Grants and funding
- R35GM131767/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01DE028916/U.S. Department of Health & Human Services | NIH | National Institute of Dental and Craniofacial Research (NIDCR)
- R21 AI149366/AI/NIAID NIH HHS/United States
- R35 GM131767/GM/NIGMS NIH HHS/United States
- R01 DE028916/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources

