PDGFRα signaling regulates cartilage and fibrous tissue differentiation during synovial joint development
- PMID: 40301343
- PMCID: PMC12041487
- DOI: 10.1038/s41467-025-59207-1
PDGFRα signaling regulates cartilage and fibrous tissue differentiation during synovial joint development
Abstract
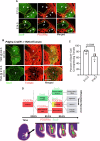
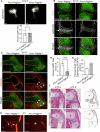
Synovial joints develop from mesenchymal structures called interzones, with progenitor cells differentiating into specialized cartilaginous and fibrous tissues of the joint. Platelet-derived growth factor receptor-α (PDGFRα) is a tyrosine kinase expressed by cells of the limb bud, but its role in limb development is unknown. To investigate PDGFRα function, we generated mice expressing mutant PDGFRα with a point mutation (D842V) that increases receptor signaling. Mutant hindlimbs are immobile with knee joints fused by cartilage and lacking ligaments and menisci. The interzone marker Gdf5 is initially expressed at E12.5 but is downregulated thereafter, suggesting a defect in interzone maintenance. Omics analysis of the joint tissues identifies ectopic cartilage matrix expressing genes for cartilage and fibrotic tissue. Thus, elevated PDGFRα signaling corrupts joint development by downregulating Gdf5 and redirecting interzone progenitors into a fibrocartilage fate. This suggests that tight regulation of tyrosine kinase activity is necessary for the development of the mouse knee joint.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Fox, A. J., Wanivenhaus, F., Burge, A. J., Warren, R. F. & Rodeo, S. A. The human meniscus: a review of anatomy, function, injury, and advances in treatment. Clin. Anat.28, 269–287 (2015). - PubMed
-
- Goldring, S. R. Pathogenesis of bone and cartilage destruction in rheumatoid arthritis. Rheumatology42, 11–16 (2003). - PubMed
-
- Lefebvre, V. & de Crombrugghe, B. Toward understanding SOX9 function in chondrocyte differentiation. Matrix Biol.16, 529–540 (1998). - PubMed
MeSH terms
Substances
Grants and funding
- R01-AR080896/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- F32-HL142222/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- F32 HL142222/HL/NHLBI NIH HHS/United States
- R01-AR073828/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- P30 GM149376/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

