Substantial spillover burden of rat hepatitis E virus in humans
- PMID: 40301345
- PMCID: PMC12041280
- DOI: 10.1038/s41467-025-59345-6
Substantial spillover burden of rat hepatitis E virus in humans
Abstract
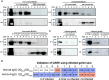
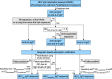
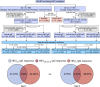
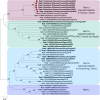
The emergence of Rocahepevirus ratti genotype 1 (rat hepatitis E virus; rat HEV) in humans presents an unprecedented threat; however, the risk of rat HEV transmission to humans is not well understood. Here, we report the "Distinguishing Antibody Response Elicitation (DARE)" method, which distinguishes exposure to rat HEV. We use four study sets from China for large-scale population analysis: set 1 (hospital visit) and set 3 (ALT abnormality) from Yunnan province, a biodiversity hotspot, and set 2 (received physical examination) and set 4 (ALT abnormality) from Jiangsu province, a non-hotspot control region. rat HEV exposure risk is significantly higher in Yunnan, with 21.97% (190 of 865) in set 1 and 13.97% (70 of 501) in set 3, compared to 0.75% (9 of 1196) in Jiangsu's set 2. Six spillover infections for rat HEV are identified in set 1, with one case of abnormal ALT. The rat-1d strains carried by rats are closely related to those human infections. Our study reveals the substantial spillover burden posed by rat HEV in biodiversity hotspots and highlights the utility of DARE method for proactive surveillance of public health emergencies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

