Biofilm inhibition of multidrug-resistant Pseudomonas aeruginosa using green-synthesized silver nanoparticles and colistin
- PMID: 40301384
- PMCID: PMC12041517
- DOI: 10.1038/s41598-025-00005-6
Biofilm inhibition of multidrug-resistant Pseudomonas aeruginosa using green-synthesized silver nanoparticles and colistin
Abstract
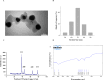
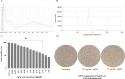
We aimed to investigate the synergistic effects of colistin and green-synthesized silver nanoparticles on the biofilm formation and expression of Quorum Sensing regulated and related genes in clinical isolates of P. aeruginosa. Ten clinical P. aeruginosa isolates collected from patients with burn wound infections were investigated. The antibiotic sensitivity pattern of the isolates was determined using disk diffusion and microbroth dilution tests. The silver nanoparticles (AgNPs) were synthesized using propolis and characterized. The microtiter plate method and scanning electron microscopy (SEM) were used to evaluate the synergistic effects of colistin and silver nanoparticles combination (AgNPs@CL) on the inhibition of biofilm formation. The effect of AgNPs@CL on the expression of genes controlled by QS was evaluated using RT-PCR. All isolates were strong biofilm formers. Confronting AgNPs@CL, all isolates were either synergistic or additive and effectively decrease the minimum inhibitory concentration (MIC) and minimum biofilm inhibitory concentration (MBIC) values of Carbapenem-Resistant P. aeruginosa (CRPA) isolates. The SEM analysis corroborated the enhanced biofilm inhibition observed with the combined treatment compared to individual AgNPs or colistin treatments. When exposed to AgNPs@CL, the expression levels of lasI, lasR, rhlI, rhlR, pelA, and pslA genes significantly decreased in P. aeruginosa ATCC 27,853 and clinical isolate No. #354, which displayed synergistic activity. In contrast, with additive activity, clinical isolate No. #30 showed no significant decrease. Targeting critical components of QS could effectively inhibit biofilm production. The results of our study suggest AgNPs@CL as an auxiliary to antibiotic therapy.
Keywords: Pseudomonas aeruginosa; Biofilms; Colistin; Drug synergism; Quorum sensing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: All experimental protocols were approved by the Ethic comity of the Hamadan University of Medical sciences, Hamadan, Iran (Ethic approval codes: IR.UMSHA.REC.1400.255). All methods were carried out in accordance with relevant guidelines and regulations. Ethical Review Board approved informed consent taken from all the participants and their legal guardians.
Figures




Similar articles
-
Synergistic effects of nano curcumin mediated photodynamic inactivation and nano-silver@colistin against Pseudomonas aeruginosa biofilms.Photodiagnosis Photodyn Ther. 2024 Feb;45:103971. doi: 10.1016/j.pdpdt.2024.103971. Epub 2024 Jan 11. Photodiagnosis Photodyn Ther. 2024. PMID: 38218569
-
Silver nanoparticle with potential antimicrobial and antibiofilm efficiency against multiple drug resistant, extensive drug resistant Pseudomonas aeruginosa clinical isolates.BMC Microbiol. 2024 Jul 26;24(1):277. doi: 10.1186/s12866-024-03397-z. BMC Microbiol. 2024. PMID: 39060955 Free PMC article.
-
Synergistic effect of silver nanoparticles and polymyxin B against biofilm produced by Pseudomonas aeruginosa isolates of pus samples in vitro.Artif Cells Nanomed Biotechnol. 2019 Dec;47(1):2465-2472. doi: 10.1080/21691401.2019.1626864. Artif Cells Nanomed Biotechnol. 2019. PMID: 31187657
-
Antibacterial and antibiofilm potential of silver nanoparticles against antibiotic-sensitive and multidrug-resistant Pseudomonas aeruginosa strains.Braz J Microbiol. 2021 Mar;52(1):267-278. doi: 10.1007/s42770-020-00406-x. Epub 2020 Nov 24. Braz J Microbiol. 2021. PMID: 33231865 Free PMC article. Review.
-
Pseudomonas aeruginosa biofilm: treatment strategies to combat infection.Arch Microbiol. 2025 May 10;207(6):141. doi: 10.1007/s00203-025-04346-8. Arch Microbiol. 2025. PMID: 40348909 Review.
Cited by
-
Biofilm-dispersal patterns in ESKAPE pathogens.Arch Microbiol. 2025 Jul 11;207(9):194. doi: 10.1007/s00203-025-04394-0. Arch Microbiol. 2025. PMID: 40643714 Review.
References
-
- WHO. Burns. Available from: https://www.who.int/news-room/fact-sheets/detail/burns (2018).
-
- Santucci, S., Gobara, S., Santos, C., Fontana, C. & Levin, A. Infections in a burn intensive care unit: Experience of seven years. J. Hosp. Infect.53, 6–13. 10.1053/jhin.2002.1340 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

