Whole-exome tumor-agnostic ctDNA analysis enhances minimal residual disease detection and reveals relapse mechanisms in localized colon cancer
- PMID: 40301653
- PMCID: PMC12202492
- DOI: 10.1038/s43018-025-00960-z
Whole-exome tumor-agnostic ctDNA analysis enhances minimal residual disease detection and reveals relapse mechanisms in localized colon cancer
Abstract
In stage 2-3 colon cancer (CC), postsurgery circulating tumor DNA (ctDNA) assessment is crucial for guiding adjuvant chemotherapy (ACT) decisions. While existing assays detect ctDNA and help identify high-risk persons with CC for recurrence, their limited sensitivity after surgery poses challenges in deciding on ACT. Additionally, a substantial portion of persons with CC fail to clear ctDNA after ACT, leading to recurrence. In this study, we performed whole-exome sequencing (WES) of ctDNA at different time points in participants with relapsed CC in two independent cohorts, alongside transcriptomic and proteomic analyses of metastases, to enhance comprehension of progression mechanisms. A plasma WES-based tumor-agnostic assay demonstrated higher sensitivity in detecting minimal residual disease (MRD) compared to current assays. Immune evasion appears to be the primary driver of progression in the localized CC setting, indicating the potential efficacy of immunotherapy for microsatellite stability in persons with CC. Organoid modeling further supports the promising potential of targeted therapy in eradicating MRD, surpassing conventional treatments.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.C. declares institutional research funding from Genentech, Merck Serono, BMS, MSD, Roche, Beigene, Bayer, Servier, Lilly, Natera, Novartis, Takeda, Astellas and Fibrogen and advisory board or speaker fees from Merck Serono, Roche, Servier, Takeda and Astellas. N.T. declares advisory board or speaker fees from Merck Serono, Servier, Pfizer, Natera and Guardant Health. M.H. declares advisory board and speaker fees from Servier. T.F. declares institutional research funding from Genentech, Adapt Immune, Roche, Beigene, Astelas, BMS, Daichii Sanyo and Amgen and speaker fees from Astrazeneca, Amgen, Bayer, BMS, Lilly, MSD and Servier. V.G. declares advisory board fees from Boehringer Ingelheim and institutional research funding from Bayer, Boehringer, Roche, Genentech, Merck Serono, Beigene, Servier, Lilly, Novartis, Takeda, Astelas, Fibrogen, Amcure, Natera, Sierra Oncology, AstraZeneca, Medimmune, BMS and MSD. S.R. declares personal fees as an invited speaker from Amgen, MSD and Servier, advisory board fees from Amgen, Servier and Sirtex and institutional funding from Ability Pharmaceuticals, Astellas, G1 Therapeutics, Hutchinson, Menarini, Mirati, Novartis, Pfizer, Pierre Fabre, Roche and Seagen. C.L.A. declares institutional research funding from Natera, C2i Genomics and BioRad Laboratories. V.P.M. reports consultancy for Johnson&Johnson and Baxter, has received honorarium for speaking at symposia and workshops by Johnson&Johnson, Medtronic and Braun Medical and has received support for attending meetings by Takeda. J. Martín-Arévalo reports consultancy for Baxter and has received honorarium for speaking at workshops by Johnson&Johnson and Medtronic. D.M. has received honorarium for speaking at symposia and workshops by Johnson&Johnson and Medtronic and support for attending meetings by Sanofi. S.G.-B., A.E. and L.P.-S. have received honorarium for educational courses by Johnson&Johnson, Marina Garcés Albir and Dixie Huntley. C.M.-C. declares advisory board or speaker fees from MSD, Astelas and BMS. The other authors declare no competing interests.
Figures
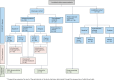



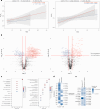
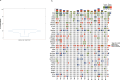

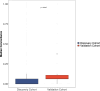





References
-
- Argilés, G. et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol.31, 1291–1305 (2020). - PubMed
-
- Tarazona, N. et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann. Oncol.30, 1804–1812 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- PI21/00689 and PI21/00695/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- FI22/00267/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- ACIF2020/381/Generalitat Valenciana (Regional Government of Valencia)
- APOSTD/2021/168/Generalitat Valenciana (Regional Government of Valencia)
- FPU21/00373/Ministerio de Educación, Cultura y Deporte (Ministry of Education, Culture and Sports, Spain)
LinkOut - more resources
Full Text Sources

