Plant development influences dynamic shifts in the root compartment microbiomes of wild and domesticated finger millet cultivars
- PMID: 40301775
- PMCID: PMC12042305
- DOI: 10.1186/s12866-025-03976-8
Plant development influences dynamic shifts in the root compartment microbiomes of wild and domesticated finger millet cultivars
Abstract
Background: Plant-microbe interactions in the rhizosphere and endosphere are crucial for maintaining plant health and ecosystem dynamics. These interactions are shaped by several factors, including the plant's developmental stage, domestication, and specific root compartments. Different plant cultivars influence unique microbial communities by secreting root exudates that either support beneficial symbionts or inhibit pathogens. This study examined the microbial community structures in the endosphere and rhizosphere of wild-type finger millet and five domesticated cultivars at two developmental stages.
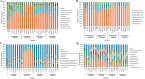
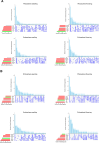
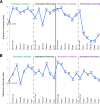
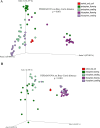
Results: Our results revealed that the plant developmental stage, root compartment, and domestication significantly influence the root-associated microbiomes. Interestingly, only about 8% of the core microbiota was consistently shared between the soil and plants, indicating that 92% shifted dynamically depending on plant type and developmental stage. Pseudomonadota, Actinomycedota, and Bacteroidota were the dominant bacterial phyla, while Ascomycota and Basidiomycota were the primary fungal phyla across all samples, displaying distinct abundance patterns. Notably, an increase in Actinomycedota in the endosphere correlated with a reduction in Pseudomonadota. The most significant shifts in microbial community composition occurred in the rhizosphere during the flowering stage, primarily driven by the genus Pseudomonas. These findings demonstrate that plant developmental stages and domestication influence the recruitment of specific microbial taxa to meet the plant's needs, particularly in various root compartments. This selective recruitment highlights the active role of plants in shaping their microbiomes, providing insights into the potential for manipulating these communities to enhance crop productivity sustainably.
Conclusion: Our results indicate that both the host developmental stage and domestication significantly influence the assembly and structure of the plant microbiome. Plant root compartments can selectively recruit specific taxa from associated core microbial communities to meet their needs, depending on the plant's developmental stage and the particular root compartment involved. These findings demonstrate that the deterministic selection pressures exerted by plants during their growth and development greatly affect their microbial communities. This has important implications for developing sustainable farming practices, reducing reliance on chemical fertilizers and pesticides, and enhancing future crop productivity.
Keywords: Amplicon sequencing; Domestication; Endosphere; Finger millet; Flowering stage; Microbiome; Plant microbiology; Plant microbiome; Rhizosphere; Seedling stage.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Plant domestication shapes rhizosphere microbiome assembly and metabolic functions.Microbiome. 2023 Mar 31;11(1):70. doi: 10.1186/s40168-023-01513-1. Microbiome. 2023. PMID: 37004105 Free PMC article.
-
Ecological Processes of Bacterial and Fungal Communities Associated with Typha orientalis Roots in Wetlands Were Distinct during Plant Development.Microbiol Spectr. 2023 Feb 14;11(1):e0505122. doi: 10.1128/spectrum.05051-22. Epub 2023 Jan 23. Microbiol Spectr. 2023. PMID: 36688664 Free PMC article.
-
Differential effects of pine wilt disease on root endosphere, rhizosphere, and soil microbiome of Korean white pine.Microbiol Spectr. 2025 Apr;13(4):e0232624. doi: 10.1128/spectrum.02326-24. Epub 2025 Mar 6. Microbiol Spectr. 2025. PMID: 40047452 Free PMC article.
-
Rhizosphere microbiome assembly, drivers and functions in perennial ligneous plant health.Microbiol Res. 2024 Oct;287:127860. doi: 10.1016/j.micres.2024.127860. Epub 2024 Jul 29. Microbiol Res. 2024. PMID: 39089083 Review.
-
Effects of Domestication on Plant-Microbiome Interactions.Plant Cell Physiol. 2022 Nov 22;63(11):1654-1666. doi: 10.1093/pcp/pcac108. Plant Cell Physiol. 2022. PMID: 35876043 Review.
References
-
- Maharajan T, Antony Ceasar S, Ajeesh Krishna TP, Ignacimuthu S. Finger millet [Eleusine coracana (L.) Gaertn]: An orphan crop with a potential to alleviate the calcium deficiency in the semi-arid tropics of Asia and Africa. Front Sustain Food Syst. 2021;5:684447.
-
- O’Kennedy MM, Grootboom A, Shewry P. Harnessing sorghum and millet biotechnology for food and health. J Cereal Sci. 2006;44(3):224–35.
-
- Wambi W, Otienno G, Tumwesigye W, Mulumba J. Genetic and genomic resources for finger millet improvement: opportunities for advancing climate-smart agriculture. J Crop Improv. 2021;35(2):204–33.
MeSH terms
LinkOut - more resources
Full Text Sources

