Molecular and clinical aspects of histone-related disorders
- PMID: 40301961
- PMCID: PMC12042324
- DOI: 10.1186/s40246-025-00734-9
Molecular and clinical aspects of histone-related disorders
Abstract
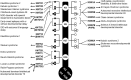
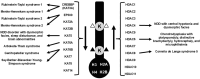
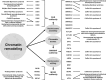
Epigenetics is the coordination of gene expression without alterations in the DNA sequence. Epigenetic gene expression is regulated by an intricate system that revolves around the interaction of histone proteins and DNA within the chromatin structure. Histones remain at the core of the epigenetic gene transcription regulation where histone proteins, along with the histone modification enzymes, and the subunits of chromatin remodelers and epigenetic readers play essential roles in regulating gene expression. Histone-related disorders encompass the syndromes induced by pathogenic variants in genes encoding histones, genes encoding histone modification enzymes, and genes encoding subunits of chromatin remodeler and epigenetic reader complexes. Defects in genes encoding histones lead to the expression of abnormal histone proteins. Abnormalities in genes encoding histone modification enzymes result in aberrant histone modifications. Defects in genes encoding subunits of the chromatin remodeler complexes result in defective chromatin remodeling. Defects in genes that code for the epigenetic readers (bromodomain proteins) will hinder their ability to regulate gene transcription. These disorders typically present manifestations that impact the nervous system which is particularly sensitive due to its need for specific patterns of gene expression for neural cell function and differentiation. To date, 72 histone-related disorders have been described including 7 syndromes due to defects in histone genes, 35 syndromes due to histone modifications defects, 26 syndromes due to defects in chromatin remodeling, and 4 due to defects in epigenetic readers. In this review article, the molecular basis of histone structure and function is first explained, followed by a summary of the histone-related syndromes.
Keywords: Bromodomain proteins; Chromatin access; Chromatin remodeling; Epigenetic readers; Histone acetylation; Histone genes; Histone methylation; Histone modification; Nucleosome assembly; Nucleosome editing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

