Red blood cell-tumor cell interactions promote tumor cell progression
- PMID: 40301999
- PMCID: PMC12042424
- DOI: 10.1186/s13046-025-03376-w
Red blood cell-tumor cell interactions promote tumor cell progression
Abstract
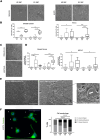
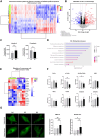
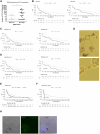
A critical step in the metastatic cascade is the survival of circulating tumor cells (CTCs) within the bloodstream. Although interactions between CTCs and various hematopoietic cells have been described, the role of red blood cells (RBCs) remains underexplored. This study investigated the interactions between tumor cells and RBCs from breast and lung cancer patients, revealing significant phenotypic and functional changes in tumor cells, unlike interactions with RBCs from healthy donors. Tumor cell and patient-derived RBC co-cultures increased tumor cell attachment and induced morphological changes. RBC-primed tumor cells showed increased adhesion, disruption of the endothelial barrier, and invasiveness, both in vitro and in vivo. Global proteome changes, including actin remodeling and VASP accumulation at cell edges, promote directional migration. RBCs from patients with metastatic breast cancer also upregulate PAK4, enhancing migration and epithelial-mesenchymal transition, whereas PAK4 inhibition reduces these effects. Clinically, a higher red blood cell distribution width (RDW) in patients with metastasis is associated with increased CTC counts and poor outcomes. This study highlights the previously unrecognized role of RBCs in promoting metastatic behavior in cancer cells and suggests potential therapeutic targets, such as PAK4, to counteract these effects.
Keywords: Actin remodeling; Breast cancer; Circulating tumor cells; Liquid biopsy; Lung cancer; Red blood cells (RBCs).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All patients and HD patients provided written informed consent. The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Galicia (2015/772) and by the Ärztekammer of Hamburg (PV-5392). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Pereira-Veiga T, Schneegans S, Pantel K, Wikman H. Circulating tumor cell-blood cell crosstalk: biology and clinical relevance. Cell Rep. 2022;40(9):111298. - PubMed
-
- Kontidou E, Collado A, Pernow J, Zhou Z. Erythrocyte-Derived MicroRNAs: emerging players in cardiovascular and metabolic disease. Arterioscler Thromb Vasc Biol. 2023;43(5):628–36. - PubMed
-
- Wojsiat J, Laskowska-Kaszub K, Mietelska-Porowska A, Wojda U. Search for Alzheimer’s disease biomarkers in blood cells: hypotheses-driven approach. Biomark Med. 2017;11(10):917–31. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

