Cytokines from parasites: manipulating host responses by molecular mimicry
- PMID: 40302223
- PMCID: PMC12203974
- DOI: 10.1042/BCJ20253061
Cytokines from parasites: manipulating host responses by molecular mimicry
Abstract
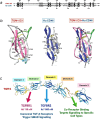
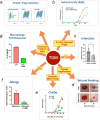
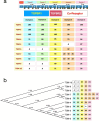
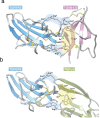
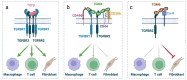
Helminth parasites have evolved sophisticated methods for manipulating the host immune response to ensure long-term survival in their chosen niche, for example, by secreting products that interfere with the host cytokine network. Studies on the secretions of Heligmosomoides polygyrus have identified a family of transforming growth factor-β (TGF-β) mimics (TGMs), which bear no primary amino acid sequence similarity to mammalian TGF-β, but functionally replicate or antagonise TGF-β effects in restricted cell types. The prototypic member, TGM1, induces in vitro differentiation of Foxp3+ T regulatory cells and attenuates airway allergic and intestinal inflammation in animal models. TGM1 is one of a family of ten TGM proteins expressed by H. polygyrus. It is a five-domain modular protein in which domains 1-2 bind TGFBR1, and domain 3 binds TGFBR2; domains 4-5 increase its potency by binding a co-receptor, CD44, highly expressed on immune cells. Domains 4-5 are more diverse in other TGMs, which bind co-receptors on cells such as fibroblasts. One variant, TGM6, lacks domains 1-2 and hence cannot transduce a signal but binds TGFBR2 through domain 3 and a co-receptor expressed on fibroblasts through domains 4-5 and blocks TGF-β signalling in fibroblasts and epithelial cells; T cells do not express the co-receptor and are not inhibited by TGM6. Hence, different family members have evolved to act as agonists or antagonists on various cell types. TGMs, which function by molecularly mimicking binding of the host cytokine to the host TGF-β receptors, are examples of highly evolved immunomodulators from parasites, including those that block interleukin (IL)-13 and IL-33 signalling, modulate macrophage and dendritic cell responses and modify host cell metabolism. The emerging panoply and potency of helminth evasion molecules illustrates the range of strategies in play to maintain long-term infections in the mammalian host.
Keywords: CD44; cytokines; evolution; immunomodulation; mimicry.
© 2025 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures
Similar articles
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Salmonella exploits host- and bacterial-derived β-alanine for replication inside host macrophages.Elife. 2025 Jun 19;13:RP103714. doi: 10.7554/eLife.103714. Elife. 2025. PMID: 40536105 Free PMC article.
-
Psychological interventions for adults who have sexually offended or are at risk of offending.Cochrane Database Syst Rev. 2012 Dec 12;12(12):CD007507. doi: 10.1002/14651858.CD007507.pub2. Cochrane Database Syst Rev. 2012. PMID: 23235646 Free PMC article.
-
TGM6 is a helminth secretory product that mimics TGF-β binding to TGFBR2 to antagonize signaling in fibroblasts.Nat Commun. 2025 Feb 21;16(1):1847. doi: 10.1038/s41467-025-56954-z. Nat Commun. 2025. PMID: 39984487 Free PMC article.
Cited by
-
Modulation of the Immune Response by Nematode Derived Molecules.Int J Mol Sci. 2025 Jun 11;26(12):5600. doi: 10.3390/ijms26125600. Int J Mol Sci. 2025. PMID: 40565065 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

