Plasmid-Mediated Co-Occurrence of mcr-1.1 in Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli Isolated From the Indigenous Seminomadic Community in Malaysia
- PMID: 40303148
- PMCID: PMC12017022
- DOI: 10.1155/2024/9223696
Plasmid-Mediated Co-Occurrence of mcr-1.1 in Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli Isolated From the Indigenous Seminomadic Community in Malaysia
Abstract
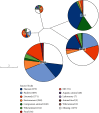
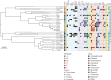
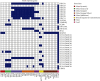
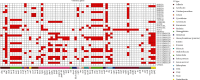
The growing prevalence of commensal antibiotic resistant Escherichia coli poses a significant concern for the global spread of antibiotic resistance. Stool samples (n = 35) from a seminomadic indigenous community in Malaysia, the Jehai, were screened for multidrug-resistant bacteria, specifically the extended-spectrum β-lactamase (ESBL) producers. Subsequently, whole-genome sequencing was used to provide genomic insights into eight ESBL-producing E. coli that colonised eight individuals. The ESBL E. coli isolates carry resistance genes from various antibiotic classes such as the β-lactams (bla TEM, bla CTX-M-15 and bla CTX-M-55), quinolones (gyrA, qnrS and qnrS1) and aminoglycosides (aph(3')-Ia, aph(6)-Id and aac(3)-IId). Three concerning convergence of ESBL, colistin and metal resistance determinants, with three plasmids from H-type lineage harbouring bla CTX-M and mcr-1.1 genes were identified. Using the Oxford Nanopore Technology (ONT) Native Barcoding Kit (SQK-NBD114.24) in conjunction with the R10.4.1 flow cell, which achieved an average read accuracy (Q > 10) of 99.84%, we further characterised the mcr-1.1-bearing plasmids, ranging in size from 25 to 28 kb, from three strains of E. coli. This report represents the first whole genome analysis of multidrug-resistant bacteria, specifically those resistant to colistin, found within the indigenous population in Malaysia. It strongly indicates that the pertinent issue of colistin resistance in the country is far more significant than previously estimated.
Keywords: antimicrobial resistance; colistin resistance; extended-spectrum β-lactamase Escherichia coli; mcr-1 gene; remote indigenous community; rural health.
Copyright © 2024 Polly Soo Xi Yap et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Emergence and Comparative Genomics Analysis of Extended-Spectrum-β-Lactamase-Producing Escherichia coli Carrying mcr-1 in Fennec Fox Imported from Sudan to China.mSphere. 2019 Nov 20;4(6):e00732-19. doi: 10.1128/mSphere.00732-19. mSphere. 2019. PMID: 31748247 Free PMC article.
-
Occurrence of the Colistin Resistance Gene mcr-1 and Additional Antibiotic Resistance Genes in ESBL/AmpC-Producing Escherichia coli from Poultry in Lebanon: A Nationwide Survey.Microbiol Spectr. 2021 Oct 31;9(2):e0002521. doi: 10.1128/Spectrum.00025-21. Epub 2021 Sep 8. Microbiol Spectr. 2021. PMID: 34494875 Free PMC article.
-
Abundance of colistin-resistant Escherichia coli harbouring mcr-1 and extended-spectrum β-lactamase-producing E. coli co-harbouring blaCTX-M-55 or -65 with blaTEM isolates from chicken meat in Vietnam.Arch Microbiol. 2022 Jan 15;204(2):137. doi: 10.1007/s00203-021-02746-0. Arch Microbiol. 2022. PMID: 35032196
-
Emerging threats: Antimicrobial resistance in extended-spectrum beta-lactamase and carbapenem-resistant Escherichia coli.Microb Pathog. 2025 Mar;200:107275. doi: 10.1016/j.micpath.2024.107275. Epub 2025 Jan 9. Microb Pathog. 2025. PMID: 39798725 Review.
-
Antimicrobial Resistance in Escherichia coli.Microbiol Spectr. 2018 Jul;6(4):10.1128/microbiolspec.arba-0026-2017. doi: 10.1128/microbiolspec.ARBA-0026-2017. Microbiol Spectr. 2018. PMID: 30003866 Free PMC article. Review.
References
-
- Loke V. P. W., Lim T., Campos-Arceiz A. Hunting Practices of the Jahai Indigenous Community in Northern Peninsular Malaysia. Global Ecology and Conservation . 2020;21 doi: 10.1016/j.gecco.2019.e00815.e00815 - DOI
-
- Mohamed-Yousif I. M., Abu J., Abdul-Aziz S., Zakaria Z., Rashid A., Awad E. A. Occurrence of Antibiotic Resistant C. jejuni and E. coli in Wild Birds, Chickens, Environment and Humans from Orang Asli Villages in Sungai Siput, Perak, Malaysia. American Journal of Animal and Veterinary Sciences . 2019;14(3):158–169. doi: 10.3844/ajavsp.2019.158.169. - DOI
-
- Mariappan V., Ngoi S. T., Lim Y. A. L., Ngui R., Chua K. H., Teh C. S. J. Genotypic and Phenotypic Characterization of Escherichia coli Isolated from Indigenous Individuals in Malaysia. Iranian Journal of Basic Medical Sciences . 2022;25(4):468–473. doi: 10.22038/IJBMS.2022.61612.13637. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

