Development and Application of an RPA-Based Rapid Point-of-Care Testing (POCT) Method for the Detection of Feline Panleukopenia Virus
- PMID: 40303162
- PMCID: PMC12016765
- DOI: 10.1155/2024/3680778
Development and Application of an RPA-Based Rapid Point-of-Care Testing (POCT) Method for the Detection of Feline Panleukopenia Virus
Abstract
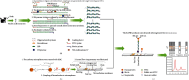
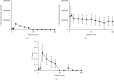
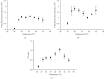
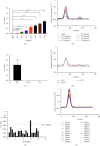
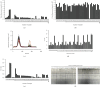
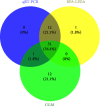
Feline panleukopenia (FP) is a highly prevalent and consequential disease that poses a substantial threat to both adult and juvenile cats across all geographical regions. The causative agent responsible for this disease is the feline panleukopenia virus (FPV). Therefore, it is imperative to develop a facile, efficient, and accurate detection method for FPV. Hence, a recombinase polymerase amplification-lateral flow dipstick assay (RPA-LFDA) method was specifically designed for the detection of FPV. The amplification process was optimized. This investigation focused on evaluating the expansion temperature detection system and revealed an optimal reaction temperature of 39°C. Then, primer combination screening involving nine groups identified F3R2 as the most effective primer set, while dilution ratio experiments determined that a 10-fold dilution yielded the best amplification products. Our findings demonstrated that the RPA-LFDA assay had an analytical sensitivity that was capable of detecting as low as 10 target copies per reaction. Furthermore, cross-reactivity tests demonstrated no interference between feline herpesvirus-1 (FHV-1) and feline calicivirus (FCV). To validate our newly developed method against existing techniques in clinical samples from three common sources on the market, we observed superior sensitivity and specificity compared to those of the colloidal gold method (CGM), with a higher positive detection rate using our nucleic acid detection system than CGM. Compared to qPCR as a reference standard, RPA-LFDA detected 39 out of 44 positive samples (including one false positive), whereas CGM detected 26 out of 44 positive samples. Based on the RPA-LFDA, the sensitivity was calculated to be 100%, the specificity was 83.33%, the mistake diagnostic rate was 16.67%, the omission diagnostic rate was 0%, and the overall accuracy reached 97.73%. Moreover, the positive coincidence rate was 97.44%, while the negative coincidence rate reached 100%. The agreement κ value was 0.8962. In conclusion, this approach exhibits greater sensitivity than CGM and offers greater convenience and cost-effectiveness than the qPCR methodology, making it a viable option for the clinical detection of FPV.
Copyright © 2024 Liang Hong et al.
Conflict of interest statement
All authors affirm that they have no conflicts of interest or competing interests associated with this study.
Figures


Similar articles
-
Development of a recombinase polymerase amplification assay with lateral flow dipstick for rapid detection of feline parvovirus.J Virol Methods. 2019 Sep;271:113679. doi: 10.1016/j.jviromet.2019.113679. Epub 2019 Jun 16. J Virol Methods. 2019. PMID: 31216435 Free PMC article.
-
The quadruplex TaqMan MGB fluorescent quantitative PCR method for simultaneous detection of feline panleukopenia virus, feline herpesvirus 1, feline calicivirus and feline infectious peritonitis virus.Front Cell Infect Microbiol. 2025 May 30;15:1581946. doi: 10.3389/fcimb.2025.1581946. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40521026 Free PMC article.
-
A novel loop-mediated isothermal amplification (LAMP) assay to diagnose feline panleukopenia.J S Afr Vet Assoc. 2024 Mar;95(1):4-10. doi: 10.36303/JSAVA.597. Epub 2024 Mar 26. J S Afr Vet Assoc. 2024. PMID: 38533815
-
Detection of protective antibody titers against feline panleukopenia virus, feline herpesvirus-1, and feline calicivirus in shelter cats using a point-of-care ELISA.J Feline Med Surg. 2011 Dec;13(12):912-8. doi: 10.1016/j.jfms.2011.07.009. Epub 2011 Aug 31. J Feline Med Surg. 2011. PMID: 21885311 Free PMC article.
-
Feline parvovirus infection and associated diseases.Vet J. 2014 Aug;201(2):150-5. doi: 10.1016/j.tvjl.2014.05.027. Epub 2014 May 22. Vet J. 2014. PMID: 24923754 Review.
References
-
- Roozitalab A., Elsakhawy O. K., Abouelkhair M. A., Matthijnssens J. M. Complete coding sequence of two feline panleukopenia virus strains isolated from domestic cats (Felis catus) in Tennessee, USA. Microbiology Resource Announcements . 2023;12(10) doi: 10.1128/MRA.00431-23.e0043123 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

