Pharmacological targeting of mitophagy via ALT001 improves herpes simplex virus 1 (HSV1)-mediated microglial inflammation and promotes amyloid β phagocytosis by restricting HSV1 infection
- PMID: 40303347
- PMCID: PMC12036882
- DOI: 10.7150/thno.105953
Pharmacological targeting of mitophagy via ALT001 improves herpes simplex virus 1 (HSV1)-mediated microglial inflammation and promotes amyloid β phagocytosis by restricting HSV1 infection
Abstract
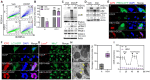
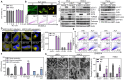
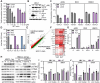
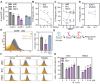
Rationale: One of the hallmarks of Alzheimer's disease (AD) is the accumulation of dysfunctional mitochondria. Herpes simplex virus type 1 (HSV1) may be a risk factor for the neuropathology linked to amyloid β (Aβ) accumulation. However, the mechanisms underlying HSV1-associated mitochondrial dysfunction in AD remain unclear. ALT001 is a novel drug that ameliorates AD-related cognitive impairment via ULK1/Rab9-mediated alternative mitophagy. In this study, we investigated the effects of ALT001 on the neurodegeneration-related microglial signatures associated with HSV1 infection. Methods: Molecular mechanisms and physiological functions of mitophagy was investigated in HSV1-infected microglia, including primary murine and human embryonic stem cell (ESC)-derived microglia (ES-MG), as well as in a microglia-neuron co-culture system. Microglial gene signatures following HSV1 infection in the presence or absence of ALT001 were analyzed using bulk RNA sequencing, and the effects of ALT001 on microglial phagocytosis and microglia-mediated immune responses were further evaluated by flow cytometry and cytokine profiles. Results: HSV1 infection inhibited PINK1/Parkin-mediated mitophagy via HSV1-encoded protein kinase US3, resulting in mitochondrial dysfunction in both human and mouse microglia. Furthermore, transcriptomic analysis of HSV1-infected microglia revealed an upregulation of distinct microglial genes associated with disease-associated microglia (DAM)-like phenotype and pro-inflammatory activity. Pharmacological targeting of mitophagy using ALT001 prevents mitochondrial damage caused by HSV1 through ULK1/Rab9-mediated pathway. Furthermore, ALT001-induced ULK1/Rab9-dependent mitophagy restricts HSV1 infection by activating interferon-mediated antiviral immunity. Consequently, ALT001 reduces HSV1-triggered neuroinflammation, recovers HSV1-altered microglial phagocytosis for Aβ, and efficiently reverses morphological and molecular abnormalities in HSV1-infected microglia by triggering mitophagy in ES-MG. ALT001 also suppressed HSV1-mediated Aβ accumulation and neurodegeneration in the microglia-neuron co-culture and cerebral organoid model. Conclusions: In this study, we identified a critical molecular link between HSV1 and AD-related microglial dysfunction. Furthermore, our findings provide an evidence that therapeutic targeting of alternative mitophagy via ALT001 effectively interfere with HSV1-induced microglial dysfunction and alleviate neurodegeneration.
Keywords: ALT001; Microglia; alternative mitophagy; herpes simplex virus 1 (HSV1); neurodegeneration.
© The author(s).
Conflict of interest statement
Competing Interests: SJO, JY, and OSS have filed a patent regarding ALT001 as an antiviral drug candidate against HSV1. JY is co-founder of Altmedical company. The other authors declare no competing interests.
Figures




References
-
- Borst K, Dumas AA, Prinz M. Microglia: Immune and non-immune functions. Immunity. 2021;54:2194–208. - PubMed
-
- Prinz M, Jung S, Priller J. Microglia Biology: One Century of Evolving Concepts. Cell. 2019;179:292–311. - PubMed
-
- Cserep C, Posfai B, Lenart N, Fekete R, Laszlo ZI, Lele Z. et al. Microglia monitor and protect neuronal function through specialized somatic purinergic junctions. Science. 2020;367:528–37. - PubMed
-
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK. et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer's Disease. Cell. 2017;169:1276–90. e17. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

