A divergent haplotype with a large deletion at the berry color locus causes a white-skinned phenotype in grapevine
- PMID: 40303437
- PMCID: PMC12038235
- DOI: 10.1093/hr/uhaf069
A divergent haplotype with a large deletion at the berry color locus causes a white-skinned phenotype in grapevine
Abstract
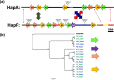
The current genetic model explaining berry skin color in Vitis vinifera is incomplete and fails to predict berry skin color phenotypes for one allele of VvMybA1, referred to as VvMybA1_SUB. Our study focuses on this specific allele, revealing that the haplotype containing VvMybA1_SUB (haplotype F) represents an ancient lineage of the berry color locus. Within haplotype F, we identified two functional subhaplotypes, HapF1 and HapF2, associated with black-skinned phenotype, and one non-functional subhaplotype, HapFDEL, responsible for white-skinned phenotype. HapF1 likely originated from wild populations domesticated in the Near East and subsequently spread globally with the expansion of viticulture. In contrast, HapF2 has a more restricted distribution and may have emerged from hybridization events between cultivated grapevines and local wild populations as viticulture migrated to the Italian peninsula. Furthermore, we found that in white-skinned berry cultivar, HapF has undergone a large deletion at the berry color locus, removing the majority of the VvMybA genes. Previous works suggested a single common origin for white-skinned varieties during grapevine domestication. Our results challenge this notion, instead proposing that white-skinned grape cultivars arose at least twice during grapevine domestication history. Alongside the major haplotype A, some white-skinned cultivars, such as cv. ‘Sultanina’ harbor HapFDEL. Since HapFDEL is present only in table grape varieties, we suggest that it likely arose from a recent mutational event and dispersed along the ancient Silk Road into East Asia. These findings enhance our understanding of the genetic diversity and evolutionary trajectory of grapevine cultivars, offering insights into their domestication and spread across different geographical regions.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Vogt T. Phenylpropanoid biosynthesis. Mol Plant. 2010;3:2–20 - PubMed
-
- Slinkard K, Singleton V. Phenol content of grape skins and the loss of ability to make anthocyanins by mutation. Vitis. 1984;23:173–5
-
- Guasch-Jané MR, Andrés-Lacueva C, Jáuregui O. et al. First evidence of white wine in ancient Egypt from Tutankhamun's tomb. J Archaeol Sci. 2006;33:1075–80
-
- Levadoux L. La sélection et l'hybridation Chez la Vigne. Montpellier: Imprimerie Charles Déhan; 1951:
LinkOut - more resources
Full Text Sources

