Comprehensive Characterization of the Oxidative Stress Profiles in Neonatal Necrotizing Enterocolitis
- PMID: 40303487
- PMCID: PMC12035842
- DOI: 10.7150/ijms.109008
Comprehensive Characterization of the Oxidative Stress Profiles in Neonatal Necrotizing Enterocolitis
Abstract
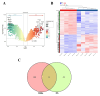
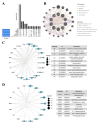
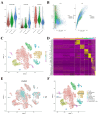
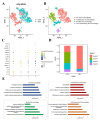
Objective: This study aims to portray the characteristics of oxidative stress (OS) in cases of Necrotizing enterocolitis (NEC), identify the hub genes and associated mechanisms involved, and explore potential drugs for NEC. Methods: We performed a comprehensive analysis integrating bulk-RNA sequencing and single-cell RNA sequencing datasets, coupled with various techniques including differential analysis, gene set enrichment analysis, and immune infiltration analysis. We aimed to systematically elucidate the variations in functions related to OS among distinct cell populations within both NEC and non-NEC tissues. Additionally, we depicted the longitudinal changes in immune cells, with a particular focus on macrophages, throughout the progression of NEC. NEC mice model was established and RT-qPCR was performed to validate the expression of the hub genes. Results: In total, 465 OS related genes were found, and 53 of them were significantly differentially expressed. These genes were mainly involved in several signaling pathways, such as TNF signaling pathway, IL-17 signaling pathway, FOXO signaling pathway, inflammatory bowel disease. The top 10 hub genes were MMP2, IL1A, MMP3, HGF, HP, IL10, PPARGC1A, TLR4, MMP9 and HMOX1. Ten kinds of drug were discovered as the potential treatment for NEC. Four specific macrophages subtypes and relative function were identified in NEC. RT-qPCR and immunofluorescence staining confirmed the expression of the hub genes in NEC model. Conclusions: This investigation yielded innovative insights into the immune environment and therapeutic methodologies directed at oxidative stress in the pathogenesis of NEC.
Keywords: immune infiltration; macrophages; neonatal necrotizing enterocolitis; oxidative stress; single cell RNA-sequencing..
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

Similar articles
-
Identification and Exploration of Pyroptosis-Related Genes in Macrophage Cells Reveal Necrotizing Enterocolitis Heterogeneity Through Single-Cell and Bulk-Sequencing.Int J Mol Sci. 2025 Apr 24;26(9):4036. doi: 10.3390/ijms26094036. Int J Mol Sci. 2025. PMID: 40362275 Free PMC article.
-
Identification of Inflammatory Genes, Pathways, and Immune Cells in Necrotizing Enterocolitis of Preterm Infant by Bioinformatics Approaches.Biomed Res Int. 2021 Apr 6;2021:5568724. doi: 10.1155/2021/5568724. eCollection 2021. Biomed Res Int. 2021. PMID: 33880370 Free PMC article.
-
miR‑34a increases inflammation and oxidative stress levels in patients with necrotizing enterocolitis by downregulating SIRT1 expression.Mol Med Rep. 2021 Sep;24(3):664. doi: 10.3892/mmr.2021.12303. Epub 2021 Jul 23. Mol Med Rep. 2021. PMID: 34296298
-
FERROPTOSIS AS A NOVEL PATHWAY IN THE PATHOGENESIS OF NECROTIZING ENTEROCOLITIS.Shock. 2025 Jul 1;64(1):12-18. doi: 10.1097/SHK.0000000000002592. Shock. 2025. PMID: 40138732 Review.
-
Oxidative Stress and Necrotizing Enterocolitis: Pathogenetic Mechanisms, Opportunities for Intervention, and Role of Human Milk.Oxid Med Cell Longev. 2018 Jul 2;2018:7397659. doi: 10.1155/2018/7397659. eCollection 2018. Oxid Med Cell Longev. 2018. PMID: 30057683 Free PMC article. Review.
References
-
- Hackam DJ, Sodhi CP. Bench to bedside - new insights into the pathogenesis of necrotizing enterocolitis. Nature reviews Gastroenterology & hepatology. 2022;19:468–79. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

