Analysis of the Relationship Between NF-κB1 and Cytokine Gene Expression in Hematological Malignancy: Leveraging Explained Artificial Intelligence and Machine Learning for Small Dataset Insights
- PMID: 40303498
- PMCID: PMC12035828
- DOI: 10.7150/ijms.109493
Analysis of the Relationship Between NF-κB1 and Cytokine Gene Expression in Hematological Malignancy: Leveraging Explained Artificial Intelligence and Machine Learning for Small Dataset Insights
Abstract
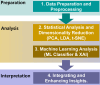
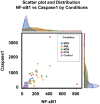
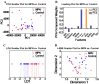
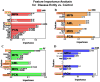
This study measures expression of nuclear factor kappa B (NF-κB)1 and related cytokine genes in bone marrow mononuclear cells in patients with hematological malignancies, analyzing the relationship between them with an integrated framework of statistical analyses, machine learning (ML), and explainable artificial intelligence (XAI). While traditional dimensionality reduction techniques-such as principal component analysis, linear discriminant analysis, and t-distributed stochastic neighbor embedding-showed limited differentiation embedding, ML classifiers (k-Nearest Neighbors, Naïve Bayes Classifier, Random Forest, and XGBoost) successfully identified critical patterns. Notably, normalized caspase-1 counts consistently emerged as the most influential feature associated with NF-κB1 activity across disease groups, as highlighted by SHapley Additive exPlanations analyses. Systematic evaluation of ML performance on small datasets revealed that a minimum sample size of 15-24 is necessary for reliable classification outcomes, particularly in cohorts of acute myeloid leukemia and myelodysplastic syndrome. These findings underscore the pivotal role of caspase-1 to the NF-κB1 gene expression in hematologic malignancy diseases. Furthermore, this study demonstrates the feasibility of leveraging ML and XAI to derive meaningful insights from limited data, offering a robust strategy for biomarker discovery and precision medicine in rare hematological malignancies.
Keywords: NF-κB / Hematological Malignancy / Machine Learning Classifiers / Explainable Artificial Intelligence / Small Data Adaptation.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
-
- Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S. et al. NF-κB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–6. - PubMed
-
- Beg A, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science274: 782-784. CrossRef CAS. 1996;274:782–4. - PubMed
-
- Grigoriadis G, Zhan Y, Grumont RJ, Metcalf D, Handman E, Cheers C. et al. The Rel subunit of NF-kappaB-like transcription factors is a positive and negative regulator of macrophage gene expression: distinct roles for Rel in different macrophage populations. The EMBO journal. 1996;15:7099–107. - PMC - PubMed
-
- Gilmore TD. Introduction to NF-κB: players, pathways, perspectives. Oncogene. 2006;25:6680–4. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

