Neuroimaging-based data-driven subtypes of spatiotemporal atrophy due to Parkinson's disease
- PMID: 40303603
- PMCID: PMC12037470
- DOI: 10.1093/braincomms/fcaf146
Neuroimaging-based data-driven subtypes of spatiotemporal atrophy due to Parkinson's disease
Abstract
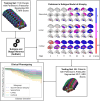
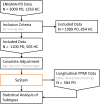
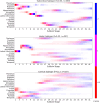
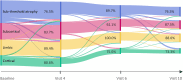
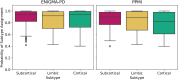
Parkinson's disease is the second most common neurodegenerative disease. Despite this, there are no robust biomarkers to predict progression, and understanding of disease mechanisms is limited. We used the Subtype and Stage Inference algorithm to characterize Parkinson's disease heterogeneity in terms of spatiotemporal subtypes of macroscopic atrophy detectable on T1-weighted MRI-a successful approach used in other neurodegenerative diseases. We trained the model on covariate-adjusted cortical thicknesses and subcortical volumes from the largest known T1-weighted MRI dataset in Parkinson's disease, Enhancing Neuroimaging through Meta-Analysis consortium Parkinson's Disease dataset (n = 1100 cases). We tested the model by analyzing clinical progression over up to 9 years in openly-available data from people with Parkinson's disease from the Parkinson's Progression Markers Initiative (n = 584 cases). Under cross-validation, our analysis supported three spatiotemporal atrophy subtypes, named for the location of the earliest affected regions as: 'Subcortical' (n = 359, 33%), 'Limbic' (n = 237, 22%) and 'Cortical' (n = 187, 17%). A fourth subgroup having sub-threshold/no atrophy was named 'Sub-threshold atrophy' (n = 317, 29%). Statistical differences in clinical scores existed between the no-atrophy subgroup and the atrophy subtypes, but not among the atrophy subtypes. This suggests that the prime T1-weighted MRI delineator of clinical differences in Parkinson's disease is atrophy severity, rather than atrophy location. Future work on unravelling the biological and clinical heterogeneity of Parkinson's disease should leverage more sensitive neuroimaging modalities and multimodal data.
Keywords: ENIGMA-PD; PPMI; clustering; disease progression modelling; neurodegeneration.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
R.S.W. has received speaking and writing honoraria from GE Healthcare, Bial, Omnix pharma and Britannia, and consultancy fees from Therakind and Accenture. N.P.O. is a paid consultant of Queen Square Analytics Limited (UK) on unrelated projects. M.T.M.H. received funding/grant support from Parkinson’s UK, Oxford NIHR BRC, University of Oxford, CPT, Lab10X, NIHR, Michael J Fox Foundation, European Platform for Neurodegenerative Disorders (EPND; H2020), GE Healthcare and the PSP Association. She also received payment for Advisory Board attendance/consultancy for Lundbeck, ESCAPE Bio, Evidera, Manus Neurodynamica, Biogen MA, CuraSen Therapeutics, Roche Products Ltd, JAZZ Pharma, Aventis Pharma. She is an advisory founder of NeuHealth Digital Ltd (company number: 14492037), a digital biomarker platform to remotely manage condition progression for Parkinson’s. The other authors report no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
