A Nomogram for Predicting Survival in Patients with SARS-CoV-2 Omicron Variant Pneumonia Based on Admission Data
- PMID: 40303607
- PMCID: PMC12039831
- DOI: 10.2147/IDR.S509178
A Nomogram for Predicting Survival in Patients with SARS-CoV-2 Omicron Variant Pneumonia Based on Admission Data
Abstract
Purpose: Patients with severe SARS-CoV-2 omicron variant pneumonia pose a serious challenge. This study aimed to develop a nomogram for predicting survival using chest computed tomography (CT) imaging features and laboratory test results based on admission data.
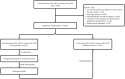
Patients and methods: A total of 436 patients with SARS-CoV-2 pneumonia (323 and 113 in the training and validation groups, respectively) were enrolled. Pneumonitis volume, assessed on chest CT scans at admission, was used to identify low- and high-risk groups. Risk analysis was performed using clinical symptoms, laboratory findings, and chest CT imaging features. A predictive algorithm was developed using Cox multivariate analysis.
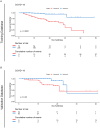
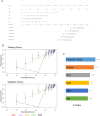
Results: The high-risk group had a shorter survival duration than the low-risk group. Significant differences in mortality rate, neutrophil and lymphocyte counts, C-reactive protein (CRP) concentration, and urea nitrogen level were observed between the two groups. In the training group, age, pneumonia volume, total bilirubin, and blood urea nitrogen were independent prognostic factors. In the validation group, age, pneumonia volume, neutrophil count, and CRP were independent prognostic factors. A personalized prediction model for survival outcomes was developed using independent predictors.
Conclusion: A personalized prediction model was created to forecast the 5-, 10-, 15-, 20-, and 30-day survival rates of patients with COVID-19 omicron variant pneumonia based on admission data, and can be used to determine the survival rate and early treatment of severe patients.
Keywords: COVID-19; omicron; pneumonia; predictive nomogram; prognosis.
© 2025 Yang et al.
Conflict of interest statement
The authors declared no potential conflicts of interest with respect to the research, authorship, and publication of this article.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

