Deciphering the Hantavirus Host Range Combining Virology and Species Distribution Models with an Emphasis on the Yellow Pygmy Rice Rat (Oligoryzomys flavescens)
- PMID: 40303704
- PMCID: PMC12017062
- DOI: 10.1155/2023/2730050
Deciphering the Hantavirus Host Range Combining Virology and Species Distribution Models with an Emphasis on the Yellow Pygmy Rice Rat (Oligoryzomys flavescens)
Abstract
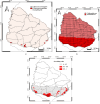
Hantaviruses are the causative agents of hantavirus pulmonary syndrome (HPS) in the Americas. In Central and South America, 28 hantavirus lineages were associated with different Sigmodontinae rodents. Of these, Lechiguanas hantavirus was initially described as a lineage associated with HPS cases in the central region of Argentina. Initial studies on the rodent hosts and viral lineages performed between 1999 and 2005 showed that HPS cases in Uruguay were distributed mostly in the southern region of the country, and that the Lechiguanas hantavirus (LECV) and the closely related Andes Central Plata hantaviruses were the viral lineages most frequently associated with HPS cases, both carried by the yellow pygmy rice rat (Oligoryzomys flavescens). Although these rodents are present all across the Uruguayan territory, determining the extent of the risk areas for hantavirus transmission based on the distribution of the infected rodents may be a useful tool for disease control and prevention. Distribution models are positioned as an effective instrument in the prediction of diseases affecting human health. Assessment of the potential distribution of rodent reservoir hosts and analysis of the influence of environmental factors on hantavirus transmission can help to understand the spatial patterns of disease transmission risk. In the present study, virological studies and species distribution models were integrated to understand the hantavirus infection risk pattern in Uruguay. Virological analyses confirmed that in Uruguay, the primary hantavirus reservoir host for both viral lineages is the yellow pygmy rice rat. Additionally, we report an Azara's grass mouse (Akodon azarae) infected with the Andes Central Plata viral lineage. Based on the seropositive and nonseropositive yellow pygmy rice rats tested, the distribution models emphasized that favorable environmental conditions for the infected rodents are mainly related to the availability of human-disturbed rural environments with high humidity. We conclude that the innovative application of the methodologies reported herein allowed for the assessment of the current risk territory for HPS in Uruguay.
Copyright © 2023 Andrés Cabrera et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
References
-
- Briese T., Alkhovsky S., Beer M., Calisher C., Charrel R., Al E. Create a new order, Bunyavirales, to accommodate nine families (eight new, one renamed) comprising thirteen genera. doi: 10.13140/RG.2.2.27230.23368. No. 2016.030a-vM, 2016. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

