High-Throughput Tiling of Essential mRNAs Increases Potency of Antisense Antibiotics
- PMID: 40304263
- PMCID: PMC12302540
- DOI: 10.1002/advs.202504284
High-Throughput Tiling of Essential mRNAs Increases Potency of Antisense Antibiotics
Abstract
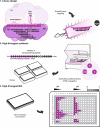
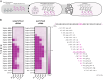
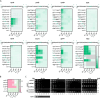
Antimicrobial resistance is outpacing drug discovery, creating an urgent need for precision-based strategies to counteract resistant pathogens. Peptide nucleic acid (PNA)-based antisense molecules offer a promising approach by selectively inhibiting essential bacterial mRNAs, but their design rules for optimal efficacy remain incompletely understood. Here, a scalable high-throughput platform is developed for the nanomolar-scale one-shot synthesis of PNAs as carrier peptide conjugates (PPNAs). Parallel synthesis of up to 1,536 PPNAs composed of up to 21 PNA or peptide building blocks enabled systematic, base-by-base analysis of RNA hybridization, mRNA inhibition, and antimicrobial activity across nine essential genes in uropathogenic Escherichia coli. The accuracy and robustness of this high-throughput tiling platform are demonstrated through in-depth analysis of the acpP mRNA and identify potent antisense inhibitors of rpsH, ftsZ, and murA. This approach provides an efficient and scalable route to design and optimize PNA-based antimicrobials, facilitating empirical testing across diverse bacterial targets. By enabling large-scale exploration of the relevant mRNA sequence space, the sequence tiling platform accelerates the discovery of antisense-based antimicrobials, offering a scalable strategy to develop precision therapies against various pathogens and combat resistance.
Keywords: Antisense Antibiotics; Asobiotics; PNA; acpP; ftsZ; mRNA; murA; rpsH.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
