Azithromycin represses evolution of ceftazidime/avibactam resistance by translational repression of rpoS in Pseudomonas aeruginosa
- PMID: 40304512
- PMCID: PMC12096824
- DOI: 10.1128/jb.00552-24
Azithromycin represses evolution of ceftazidime/avibactam resistance by translational repression of rpoS in Pseudomonas aeruginosa
Abstract
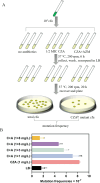
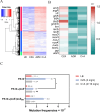
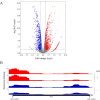
Antibiotic combinations can slow down resistance development and/or achieve synergistic therapeutic effects. In this study, we observed that a combined use of ceftazidime-avibactam (CZA) with azithromycin effectively repressed CZA resistance development in Pseudomonas aeruginosa. Transcriptome analysis revealed that subinhibitory concentrations of azithromycin reduced the expression of genes involved in stress-induced mutagenesis, including the stress response sigma factor rpoS. Interestingly, ribosome profiling revealed global redistribution of ribosomes by azithromycin, among which ribosome stalling was significantly intensified near the 5´ terminus of the rpoS mRNA. Further DNA mutational analysis revealed that azithromycin represses the translation of rpoS through its 5´-terminal rare codons, which in turn reduced its transcription. These in vitro observations have been recapitulated in vivo where azithromycin-repressed CZA resistance development when P. aeruginosa was passaged in mice. Overall, our study revealed the molecular mechanism of azithromycin-mediated repression of antibiotic resistance development, providing a promising antibiotic combination for the treatment of P. aeruginosa infections.IMPORTANCEAntibiotic resistance, a global public health challenge, demands the development of novel antibiotics and therapeutic strategies. Ceftazidime-avibactam (CZA) is a combination of a β-lactam antibiotic with a β-lactamase inhibitor that is effective against various gram-negative bacteria such as Pseudomonas aeruginosa. However, clinical CZA-resistant isolates have been reported. Here, we found that combining CZA with azithromycin can effectively suppress the development of resistance in P. aeruginosa in vitro and in vivo. Moreover, we found that azithromycin represses the translation initiation of rpoS through its 5´-terminal rare and less frequent codons, thereby subsequently reducing the mutational frequency of CZA resistance. Therefore, our work provides a promising antibiotic combination for the treatment of P. aeruginosa infections.
Keywords: Pseudomonas aeruginosa; antibiotic combination; ceftazidime-avibactam.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
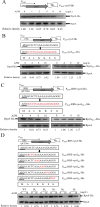

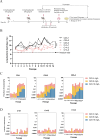
References
-
- Uddin TM, Chakraborty AJ, Khusro A, Zidan BRM, Mitra S, Emran TB, Dhama K, Ripon MKH, Gajdács M, Sahibzada MUK, Hossain MJ, Koirala N. 2021. Antibiotic resistance in microbes: history, mechanisms, therapeutic strategies and future prospects. J Infect Public Health 14:1750–1766. doi:10.1016/j.jiph.2021.10.020 - DOI - PubMed
-
- WHO . 2024. Bacterial Priority Pathogens List, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. Geneva: World Health Organization; Licence: CC BY-NC-SA 30 IGO
-
- Berrazeg M, Jeannot K, Ntsogo Enguéné VY, Broutin I, Loeffert S, Fournier D, Plésiat P. 2015. Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 59:6248–6255. doi:10.1128/AAC.00825-15 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

