Altered miRNA expression in duodenal tissue of celiac patients and the impact of a gluten-free diet: a preliminary study
- PMID: 40304865
- PMCID: PMC12043776
- DOI: 10.1007/s11033-025-10534-y
Altered miRNA expression in duodenal tissue of celiac patients and the impact of a gluten-free diet: a preliminary study
Abstract
Background: MicroRNAs (miRNAs) are crucial regulators of gene expression, impacting a wide range of biological processes. Their dysregulation can result in pathological changes and contribute to the development of various disorders. This study aims to evaluate the expression of selected miRNAs in duodenal tissue of paediatric patients with active celiac disease (CD), investigate the role of dysregulated miRNAs in disease pathogenesis and assess the changes in their expression profile in response to a gluten-free diet (GFD).
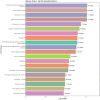
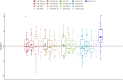
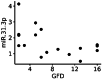
Methods and results: The study included newly diagnosed celiac patients (n = 20), celiac patients adhering to a GFD (n = 17) and a control group (n = 29). The miRNA expression in duodenal samples was quantified by real-time PCR. Dysregulated miRNAs were analysed for functional enrichment in molecular pathways. Our results identified 8 dysregulated miRNAs in celiac patients: miR-155-5p (upregulated) and hsa-miR-22-5p, hsa-miR-192-5p, hsa-miR-338-3p, hsa-miR-31-5p, hsa-miR-31-3p, hsa-miR-215-5p and hsa-miR-378d (downregulated). Pathway analysis implicated these miRNAs in regulating various signaling pathways related to inflammation, immune response and intercellular junctions, all of which are relevant to the pathogenesis of CD. Moreover, miR-31-3p was upregulated in CD patients on a GFD, exhibiting a negative correlation with the duration of GFD. For other miRNAs, the level of expression in CD patients adhering to a GFD was restored to levels similar to those observed in the control group.
Conclusion: This preliminary study reveals significant changes in miRNA expression in duodenal biopsies from paediatric CD patients and how these patterns shift with dietary intervention. Understanding the interactions among dysregulated miRNAs may lead to novel therapeutic strategies for managing CD.
Keywords: Celiac disease; Dysregulation; Gluten-free diet; MiRNA; Molecular pathways.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethical commitee of Jessenius Faculty of Medicine in Martin, Comenius University in Bratislava (EK 60/2018). Consent to participate: Informed consent was obtained from all individual participants/their legal guardians included in the study. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Circulating miRNAs as Potential Biomarkers for Celiac Disease Development.Front Immunol. 2021 Dec 7;12:734763. doi: 10.3389/fimmu.2021.734763. eCollection 2021. Front Immunol. 2021. PMID: 34950132 Free PMC article.
-
miRNAs affect the expression of innate and adaptive immunity proteins in celiac disease.Am J Gastroenterol. 2014 Oct;109(10):1662-74. doi: 10.1038/ajg.2014.203. Epub 2014 Jul 29. Am J Gastroenterol. 2014. PMID: 25070052
-
The role of mir-197-3p in regulating the tight junction permeability of celiac disease patients under gluten free diet.Mol Biol Rep. 2023 Mar;50(3):2007-2014. doi: 10.1007/s11033-022-08147-w. Epub 2022 Dec 19. Mol Biol Rep. 2023. PMID: 36536183
-
Expression of MicroRNAs in Adults with Celiac Disease: A Narrative Review.Int J Mol Sci. 2024 Aug 30;25(17):9412. doi: 10.3390/ijms25179412. Int J Mol Sci. 2024. PMID: 39273359 Free PMC article. Review.
-
Salivary and fecal microbiota and metabolome of celiac children under gluten-free diet.Int J Food Microbiol. 2016 Dec 19;239:125-132. doi: 10.1016/j.ijfoodmicro.2016.07.025. Epub 2016 Jul 19. Int J Food Microbiol. 2016. PMID: 27452636 Review.
References
-
- Gatti S, Rubio-Tapia A, Makharia G, Catassi C (2024) Patient and community health global burden in a world with more Celiac disease. Gastroenterology 167:23–33. 10.1053/j.gastro.2024.01.035 - PubMed
-
- Hunt KA, van Heel DA (2009) Recent advances in coeliac disease genetics. Gut 58:473–476. 10.1136/gut.2008.155879 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

