The Glutamate-gated Chloride Channel Facilitates Sleep by Enhancing the Excitability of Two Pairs of Neurons in the Ventral Nerve Cord of Drosophila
- PMID: 40304877
- PMCID: PMC12494514
- DOI: 10.1007/s12264-025-01397-1
The Glutamate-gated Chloride Channel Facilitates Sleep by Enhancing the Excitability of Two Pairs of Neurons in the Ventral Nerve Cord of Drosophila
Abstract
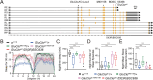
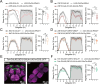
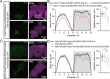
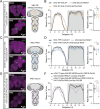
Sleep, an essential and evolutionarily conserved behavior, is regulated by numerous neurotransmitter systems. In mammals, glutamate serves as the wake-promoting signaling agent, whereas in Drosophila, it functions as the sleep-promoting signal. However, the precise molecular and cellular mechanisms through which glutamate promotes sleep remain elusive. Our study reveals that disruption of glutamate signaling significantly diminishes nocturnal sleep, and a neural cell-specific knockdown of the glutamate-gated chloride channel (GluClα) markedly reduces nocturnal sleep. We identified two pairs of neurons in the ventral nerve cord (VNC) that receive glutamate signaling input, and the GluClα derived from these neurons is crucial for sleep promotion. Furthermore, we demonstrated that GluClα mediates the glutamate-gated inhibitory input to these VNC neurons, thereby promoting sleep. Our findings elucidate that GluClα enhances nocturnal sleep by mediating the glutamate-gated inhibitory input to two pairs of VNC neurons, providing insights into the mechanism of sleep promotion in Drosophila.
Keywords: Drosophila; Glutamate-gated chloride channel; Neural activity; Neural circuit; Sleep; Ventral nerve cord.
© 2025. The Author(s).
Conflict of interest statement
Conflict of interest: The authors declare no competing financial interests.
Figures


References
-
- Nitz DA, van Swinderen B, Tononi G, Greenspan RJ. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr Biol 2002, 12: 1934–1940. - PubMed
-
- van Swinderen B, Nitz DA, Greenspan RJ. Uncoupling of brain activity from movement defines arousal States in Drosophila. Curr Biol 2004, 14: 81–87. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

