VAC14 oligomerization is essential for the function of the FAB1/PIKfyve-VAC14-FIG4 complex
- PMID: 40305106
- PMCID: PMC12260172
- DOI: 10.1091/mbc.E24-11-0490
VAC14 oligomerization is essential for the function of the FAB1/PIKfyve-VAC14-FIG4 complex
Abstract
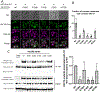
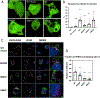
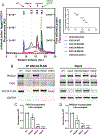
The PIKfyve-VAC14-FIG4 complex synthesizes and turns over phosphatidylinositol-3,5-bisphosphate, PI(3,5)P2, an essential signaling lipid. A medium-resolution structure revealed that VAC14 forms a star-shaped pentamer scaffold. Two legs of VAC14 bind FIG4, with one leg also occupied by PIKfyve. The significance of VAC14 oligomerization was unknown. Here, using Alphafold2 and cryogenic electron microscopy maps we generated an atomic-resolution prediction, and found that some mutations linked to pediatric neurodegenerative diseases reside in the VAC14-VAC14 interfaces. A corresponding yeast mutation, along with additional mutations, demonstrates that VAC14 oligomerization is critical for Fab1/PIKfyve function. These mutations cause defects in the generation of PI(3,5)P2, in VAC14 localization, and in VAC14 oligomerization. Similarly, VAC14 patient mutations expressed in human VAC14 knockout (KO) cells, are defective in the formation of the PIKfyve-VAC14-FIG4 complex, as measured by pull-down assays, are defective in VAC14 oligomerization as measured by fluorescence-detection size-exclusion chromatography of cell lysates, and are defective in colocalization with VPS35-containing endosomes. These studies show that VAC14 oligomerization plays a crucial role in the regulation of PIKfyve/FAB1 and provide insights into selected patient mutations. Moreover, they suggest that small molecules that stabilize the VAC14 complex may provide an intervention for diseases linked to mutations in VAC14.
Conflict of interest statement
Conflicts of interest: The authors declare no financial conflict of interest.
Figures
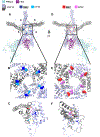


References
-
- Cianfrocco MA, Wong-Barnum M, Youn C, Wagner R, and Leschziner A (2017). COSMIC2: A science gateway for cryo-electron microscopy structure determination. In: ACM International Conference Proceeding Series.
-
- Cooke FT, Dove SK, McEwen RK, Painter G, Holmes AB, Hall MN, Michell RH, and Parker PJ (1998). The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr Biol 8. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

