Multiple association studies identify 3 novel candidate genes for teat number trait in Danish Landrace and Large White pigs: BRINP3, LIN52, and UBE3B
- PMID: 40305433
- PMCID: PMC12124257
- DOI: 10.1093/jas/skaf145
Multiple association studies identify 3 novel candidate genes for teat number trait in Danish Landrace and Large White pigs: BRINP3, LIN52, and UBE3B
Abstract
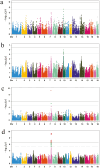
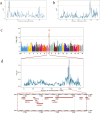
Milk is an essential source of nutrition for preweaning piglets. Therefore, in the breeding process, sows were expected to have sufficient teats to suckle their piglets. However, in Danish Landrace and Large White pigs, the number of piglets born currently exceeds the number of teats, making it urgent to select and breed for an increased teat number. In this study, the samples of 491 Danish Landrace pigs and 1,047 Danish Large White pigs with teat number phenotype were used to perform genome-wide association studies to identify SNPs associated with total teat number (TTN) based on SNP-chip data and data imputed to the level of whole-genome sequencing (iWGS), respectively. In Landrace pigs, the most significant SNP on SSC10 explains 5.14% of the phenotypic variance, while in Large White pigs, the most significant SNP on SSC7 explains 4.46% of the phenotypic variance. Additionally, linkage disequilibrium and linkage analysis (LDLA) were used to refine the regions of QTLs on SSC10 to 2.89 to 5.43 Mb in Danish Landrace pigs and to 96.00 to 97.95 Mb on SSC7 in Danish Large White pigs, respectively. To maximize the utility of information from 2 populations, meta-analysis was conducted across multiple populations. A total of 12 protein-coding genes were identified within the candidate QTL regions determined by LDLA and meta-analysis. To supplement the candidate gene set, transcriptome-wide association studies (TWAS) based on embryo and placenta tissues identified 7 protein-coding genes associated with TTN in Landrace and Large White pigs. Phenome-wide association studies (PheWAS) query was conducted for all the above genes, revealing that nearly all of them are associated with teat number traits. Additionally, some genes showed strong associations with carcass traits, suggesting a potential association between teat number and carcass traits. Through functional annotation and integrated analysis, BRINP3, LIN52, ABCD4, and UBE3B were determined as the functional candidate genes regulating TTN. These findings lay the foundation for identifying the genetic loci regulating teat number in Danish pigs, as well as for their molecular breeding.
Keywords: candidate gene; genome-wide association study; pig; teat number.
Plain language summary
Piglets depend on milk of sows for survival, making it crucial for sows to have enough teats to feed them. However, in Danish Landrace and Large White pigs, the number of piglets often exceeds the number of teats. In this study, the samples of 491 Danish Landrace pigs and 1,047 Danish Large White pigs were performed genome-wide associate studies to identify genetic loci influencing teat number. Through multiple association analyses, BRINP3, LIN52, ABCD4, and UBE3B were identified as functional candidate genes regulating Danish Landrace and Large White pigs. Some of these genes were also found to be associated with carcass traits, suggesting that a connection between teat number and body composition exists. Valuable insights into the genetics of teat number were provided by these findings, offering important information for breeding pigs with improved maternal traits.
© The Author(s) 2025. Published by Oxford University Press on behalf of the American Society of Animal Science.
Figures




References
-
- Alexopoulos, J. G., Lines D. S., Hallett S., and Plush K. J... 2018. A review of success factors for piglet fostering in lactation. Animals (Basel). 8:38. doi: https://doi.org/10.3390/ani8030038 - DOI - PMC - PubMed
-
- Bovo, S., Ballan M., Schiavo G., Ribani A., Tinarelli S., Utzeri V. J., Dall’Olio S., Gallo M., and Fontanesi L... 2021. Single-marker and haplotype-based genome-wide association studies for the number of teats in two heavy pig breeds. Anim. Genet. 52:440–450. doi: https://doi.org/10.1111/age.13095 - DOI - PMC - PubMed
-
- Browning, S. R., and Browning B. L... 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81:1084–1097. doi: https://doi.org/10.1086/521987 - DOI - PMC - PubMed
-
- Browning, B. L., Zhou Y., and Browning S. R... 2018. A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 103:338–348. doi: https://doi.org/10.1016/j.ajhg.2018.07.015 - DOI - PMC - PubMed
-
- Chang, C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., and Lee J. J... 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 4:7. doi: https://doi.org/10.1186/s13742-015-0047-8 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

