A multimodal and fully automated system for prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer
- PMID: 40305609
- PMCID: PMC12042891
- DOI: 10.1126/sciadv.adr1576
A multimodal and fully automated system for prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer
Abstract
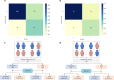
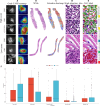
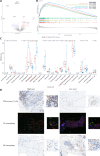
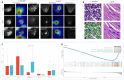
Accurately predicting pathological complete response (pCR) before neoadjuvant chemotherapy (NAC) is crucial for patients with breast cancer. In this study, we developed a multimodal integrated fully automated pipeline system (MIFAPS) in forecasting pCR to NAC, using a multicenter and prospective dataset of 1004 patients with locally advanced breast cancer, incorporating pretreatment magnetic resonance imaging, whole slide image, and clinical risk factors. The results demonstrated that MIFAPS offered a favorable predictive performance in both the pooled external test set [area under the curve (AUC) = 0.882] and the prospective test set (AUC = 0.909). In addition, MIFAPS significantly outperformed single-modality models (P < 0.05). Furthermore, the high deep learning scores were associated with immune-related pathways and the promotion of antitumor cells in the microenvironment during biological basis exploration. Overall, our study demonstrates a promising approach for improving the prediction of pCR to NAC in patients with breast cancer through the integration of multimodal data.
Figures



References
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). - PubMed
-
- Gradishar W. J., Anderson B. O., Balassanian R., Blair S. L., Burstein H. J., Cyr A., Elias A. D., Farrar W. B., Forero A., Giordano S. H., Goetz M. P., Goldstein L. J., Isakoff S. J., Lyons J., Marcom P. K., Mayer I. A., McCormick B., Moran M. S., O'Regan R. M., Patel S. A., Pierce L. J., Reed E. C., Salerno K. E., Schwartzberg L. S., Sitapati A., Smith K. L., Smith M. L., Soliman H., Somlo G., Telli M. L., Ward J. H., Kumar R., Shead D. A., Breast Cancer, version 4.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 16, 310–320 (2018). - PubMed
-
- Kuerer H. M., Smith B. D., Krishnamurthy S., Yang W. T., Valero V., Shen Y., Lin H., Lucci A., Boughey J. C., White R. L., Diego E. J., Rauch G. M., Exceptional Responders Clinical Trials Group , Eliminating breast surgery for invasive breast cancer in exceptional responders to neoadjuvant systemic therapy: A multicentre, single-arm, phase 2 trial. Lancet Oncol. 23, 1517–1524 (2022). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

