Inhibition of MBTPS1 enhances antitumor immunity and potentiates anti-PD-1 immunotherapy
- PMID: 40307212
- PMCID: PMC12043911
- DOI: 10.1038/s41467-025-59193-4
Inhibition of MBTPS1 enhances antitumor immunity and potentiates anti-PD-1 immunotherapy
Abstract
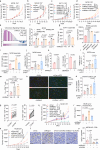
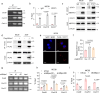
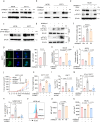
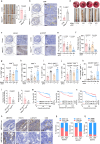
Despite advances in cancer immunotherapy, colorectal cancer patients exhibit limited therapeutic responses. Therefore, the exploration of strategies combining immunotherapy with adjuvant approaches to enhance adaptive immune responses is in demand. Here, we perform a customized in vivo CRISPR-Cas9 screen to target genes encoding membrane and secreted proteins in CRC mouse models with different immune characteristics. We observe that loss of membrane-bound transcription factor site-1 protease (MBTPS1) in tumor cells enhances antitumor immunity and potentiates anti-PD-1 therapy. Mechanistic studies reveal that tumor cell-intrinsic MBTPS1 competes with USP13 for binding to STAT1, thereby disrupting the USP13-dependent deubiquitination-mediated STAT1 stabilization. The upregulated STAT1-transcribed chemokines including CXCL9, CXCL10, and CXCL11, promote CXCR3+CD8+ T cell infiltration. Notably, the regulatory role of MBTPS1 in antitumor immunity operates independently of its classic function in cleaving membrane-bound transcription factors. Collectively, our results provide a theoretical basis for MBTPS1 as a potential immunotherapy target.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

