Identification of soluble biomarkers that associate with distinct manifestations of long COVID
- PMID: 40307449
- PMCID: PMC12043503
- DOI: 10.1038/s41590-025-02135-5
Identification of soluble biomarkers that associate with distinct manifestations of long COVID
Abstract
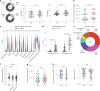
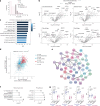
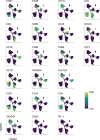
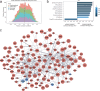
Long coronavirus disease (COVID) is a heterogeneous clinical condition of uncertain etiology triggered by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Here we used ultrasensitive approaches to profile the immune system and the plasma proteome in healthy convalescent individuals and individuals with long COVID, spanning geographically independent cohorts from Sweden and the United Kingdom. Symptomatic disease was not consistently associated with quantitative differences in immune cell lineage composition or antiviral T cell immunity. Healthy convalescent individuals nonetheless exhibited higher titers of neutralizing antibodies against SARS-CoV-2 than individuals with long COVID, and extensive phenotypic analyses revealed a subtle increase in the expression of some co-inhibitory receptors, most notably PD-1 and TIM-3, among SARS-CoV-2 nonspike-specific CD8+ T cells in individuals with long COVID. We further identified a shared plasma biomarker signature of disease linking breathlessness with apoptotic inflammatory networks centered on various proteins, including CCL3, CD40, IKBKG, IL-18 and IRAK1, and dysregulated pathways associated with cell cycle progression, lung injury and platelet activation, which could potentially inform the diagnosis and treatment of long COVID.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S. Aleman has received honoraria for educational events and lectures unrelated to this work from Gilead, AbbVie, Biogen and MSD and reports grants from Gilead and AbbVie. M.B. is a consultant for Bristol Myers Squibb, Mabtech, Pfizer, Oxford Immunotec and MSD. The other authors declare no competing interests.
Figures









References
MeSH terms
Substances
Grants and funding
- 2020-06121/Vetenskapsrådet (Swedish Research Council)
- 2021-01141/Vetenskapsrådet (Swedish Research Council)
- 2021-04779/Vetenskapsrådet (Swedish Research Council)
- 2022-01313/Vetenskapsrådet (Swedish Research Council)
- 2021-06534/Vetenskapsrådet (Swedish Research Council)
- 2021.0136/Knut och Alice Wallenbergs Stiftelse (Knut and Alice Wallenberg Foundation)
- CG-22 0009/Svenska Sällskapet för Medicinsk Forskning (Swedish Society for Medical Research)
- PG-22-0432-H-01/Svenska Sällskapet för Medicinsk Forskning (Swedish Society for Medical Research)
- 22 2237 Pj/Cancerfonden (Swedish Cancer Society)
- 2019-00969/Karolinska Institutet (Karolinska Institute)
- 2021-00513/Karolinska Institutet (Karolinska Institute)
- 2022-01719/Karolinska Institutet (Karolinska Institute)
- 2022-307/Magnus Bergvalls Stiftelse (Magnus Bergvall Foundation)
- P500PB_211069/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- COV-LT2-0041/DH | National Institute for Health Research (NIHR)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

