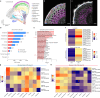
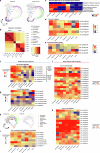
Extended Data Fig. 7. Comparative expression analysis of cell wall remodelling genes in compacted soil conditions with scRNA-seq and bulk RNA-seq approaches.
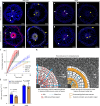
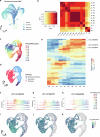
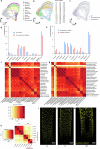
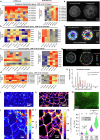
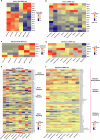
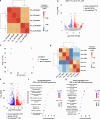
a, Left panel: Heatmap showing increased expression (log2 fold change) of xyloglucan biosynthesis genes in compacted soil conditions compared to non-compacted soil conditions in Xkitaake as detected by scRNA-seq. Xyloglucan biosynthesis genes are broadly upregulated across major cell types, with a slightly higher induction observed in the exodermis. The stronger induction of xyloglucan biosynthesis genes in the exodermis aligns with the observed barrier reinforcement. NC: Non-compacted soils. CMP: Compacted soils. Right panel: Heatmap showing scaled expression of xyloglucan biosynthesis genes in non-compacted and compacted soil conditions in Xkitaake, as revealed by bulk RNA-seq. Bulk RNA-seq also reveals a general induction of xyloglucan biosynthesis genes in compacted soils. The upregulation of these genes suggests enhanced cell wall reinforcement in response to compacted soil conditions. b, Left panel: Heatmap showing differential expression (log2 fold change) of cellulose synthase (CESA) genes in compacted soil conditions compared to non-compacted soil conditions in Xkitaake as detected by scRNA-seq. Notably, CESA4, CESA7, and CESA8 exhibit increased gene expression in sclerenchyma, suggesting enhanced secondary cell wall formation in this tissue under soil compaction. Right panel: Heatmap showing scaled expression of CESA genes in non-compacted and compacted soil conditions in Xkitaake, as revealed by bulk RNA-seq. Interestingly, bulk RNA seq reveals a general down-regulation of CESA genes in compacted soils, although the decrease is subtle for most examined genes. The relatively stronger down-regulation of CESA1, CESA5, and CESA6, combined with the relatively weaker down-regulation of CESA4, CESA7, and CESA8, may suggest a transition toward secondary cell wall deposition. c, Left panel: Heatmap showing increased expression (log2 fold change) of expansin genes in compacted soil conditions compared to non-compacted soil conditions in Xkitaake as detected by scRNA-seq. Increased expression of expansin genes, particularly in the exodermis and cortex cell layers, suggests the enhanced cell expansion at exodermis and cortex under soil compaction. Right panel: Heatmap showing scaled expression of expansin genes in non-compacted and compacted soil conditions in Xkitaake, as revealed by bulk RNA-seq. Bulk RNA-seq further supports the upregulation of expansin genes in compacted soil conditions. The up-regulation of expansin genes correlates with the radial expansion of rice roots in response to soil compaction. d, Left panel: Heatmap showing increased expression (log2 fold change) of xylanase inhibitor genes in compacted soil conditions compared to non-compacted soil conditions in Xkitaake as detected by scRNA-seq. Xylanase inhibitor genes are broadly upregulated across major cell types. As xylanase inhibitor is tightly relevant to defense response, it further suggests that soil compaction could induce defense response in rice root. Right panel: Heatmap showing scaled expression of xylanase inhibitor encoding genes in non-compacted and compacted soil conditions in Xkitaake, as revealed by bulk RNA-seq. Bulk RNA-seq analysis further supports the upregulation of xylanase inhibitor genes in compacted soil conditions. Grey boxes mean that the gene was not detected during the comparative analysis. Annotation for the included genes: LOC-Os02g57770, OsXTH22; LOC-Os03g01800, OsXTH19; LOC-Os08g24750, Xyloglucan fucosyltransferase8; LOC-Os06g10960, Xyloglucan fucosyltransferase2; LOC-Os03g13570, OsXTH28; LOC-Os09g28460, Xyloglucan fucosyltransferase7; LOC-Os10g32170, Xyloglucan galactosyltransferase KATAMARI1 homolog; LOC-Os09g20850, OsTBL41, Xyloglucan O-acetyltransferase 2; LOC-Os03g05060, Xyloglucan galactosyltransferase KATAMARI1 homolog; LOC-Os04g51510, OsXTH7. LOC_Os01g71080, xylanase inhibitor; LOC_Os01g71094, xylanase inhibitor; LOC_Os01g71130, xylanase inhibitor; LOC_Os01g71070, xylanase inhibitor; LOC_Os03g10478, endo-1,4-beta-xylanase 5-like; LOC_Os08g40690, xylanase inhibitor LOC_Os08g40680, xylanase inhibitor. The complete list of gene ID and annotations are included in Supplementary Table 14. e, Confocal imaging of root transverse sections from non-compacted and compacted soil conditions. The cell boundary was visualized by the auto-fluorescence activated by a 405 nm wavelength laser. Scale bars: 100 μm. n = 3 biological replicates (roots), all showing similar trends. f, The heatmap of cortical cell areas under both non-compacted and compacted soil conditions. Red and blue colors indicate bigger and smaller cells, respectively. n = 3 biological replicates (roots), all showing similar trends. Scale bar: 50 μm. g, The quantification of exodermal cell areas in the root transverse sections. For the heatmap, red and blue indicate bigger and smaller cells, respectively. Segmented cells are outlined in cyan and superimposed on the meshed surface where the cell wall signals are projected (greyscale). n = 3 biological replicates (roots), all showing similar trends. Scale bar: 50 μm. h, The histogram showing the cell area distribution of exodermal cell under both non-compacted and compacted soil conditions. 3 biological replicates (roots) are included. i,j, Non-compacted and compacted (respectively) endodermal region maps of the Brillouin frequency shift (relative to the shift in the cytoplasm ΔfB = 0) demonstrating apparent greater cell-wall stiffness in the compacted case. The primary roots were harvested from compacted and non-compacted soils were radially sectioned and imaged using Brillouin microscopy. k,l, Similarly, maps of acoustic attenuation between the two cases demonstrate greater apparent longitudinal viscosity in the compacted soil conditions. Scale bars in i and k: 10 μm; Scale bar in j and l: 10 μm. m, Brightfield image of a rice root radial cross-section (red boxes are the relevant regions of interest in i-l). Scale bar: 50 μm. n, Violin plot showing the cell wall stiffness of rice primary root in compacted and non-compacted soil conditions. The width of each violin represents the kernel density estimation of the data distribution. The solid line within each violin denotes the median, while the dashed lines represent the first quartile (Q1) and third quartile (Q3). Q1 corresponds to the 25th percentile of the data, and Q3 corresponds to the 75th percentile. Yuen’s t-tests (two-tailed) indicate that there is no statistically significant (NS, p value = 0.7500) relative shift in Brillouin frequency between the cytoplasm (control) regions in non-compacted (NC) and compacted (C) specimens. However, there is a clear frequency shift between endodermal cell-walls between the two cases (p value < 1.0000 × 10−10; *: p < 0.05) indicating greater elasticity for compacted cell-walls. Four cross-sections for each case of compacted and non-compacted were imaged containing 35 and 44 cells respectively. For cell wall measurements, n = 1744 (non-compaction, endodermis), 1843 (compaction, endodermis), 11852 (non-compaction, cytoplasm), and 17592 (compaction, cytoplasm) units, were analyzed respectively. Source Data