Cell cycle duration determines oncogenic transformation capacity
- PMID: 40307557
- PMCID: PMC12119354
- DOI: 10.1038/s41586-025-08935-x
Cell cycle duration determines oncogenic transformation capacity
Abstract
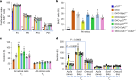
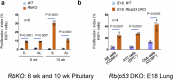
Oncogenic mutations are widespread in normal human tissues1. Similarly, in murine chimeras, cells carrying an oncogenic lesion contribute normal cells to adult tissues without causing cancer2-4. How lineages that escape cancer via normal development differ from the minority that succumb is unclear. Tumours exhibit characteristic cancer hallmarks; we therefore searched for hallmarks that differentiate cancer-prone lineages from resistant lineages. Here we show that total cell cycle duration (Tc) predicts transformation susceptibility across multiple tumour types. Cancer-prone Rb- and p107-deficient retina (Rb is also known as Rb1 and p107 is also known as Rbl1) exhibited defects in apoptosis, senescence, immune surveillance, angiogenesis, DNA repair, polarity and proliferation. Perturbing the SKP2-p27-CDK2/CDK1 axis could block cancer without affecting these hallmarks. Thus, cancer requires more than the presence of its hallmarks. Notably, every tumour-suppressive mutation that we tested increased Tc, and the Tc of the cell of origin of retinoblastoma cells was half that of resistant lineages. Tc also differentiated the cell of origin in Rb-/- pituitary cancer. In lung, loss of Rb and p53 (also known as Trp53) transforms neuroendocrine cells, whereas KrasG12D or BrafV600E mutations transform alveolar type 2 cells5-7. The shortest Tc consistently identified the cell of origin, regardless of mutation timing. Thus, relative Tc is a hallmark of initiation that distinguishes cancer-prone from cancer-resistant lineages in several settings, explaining how mutated cells escape transformation without inducing apoptosis, senescence or immune surveillance.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

