Porphyromonas gingivalis induces Zbp1-mediated macrophages PANoptosis in periodonitis pathophysiology
- PMID: 40307566
- PMCID: PMC12130536
- DOI: 10.1038/s12276-025-01443-y
Porphyromonas gingivalis induces Zbp1-mediated macrophages PANoptosis in periodonitis pathophysiology
Abstract
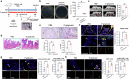
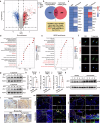
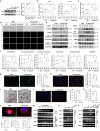
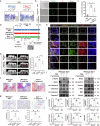
Periodontitis is an oral immunoinflammatory disease, and macrophages play a crucial role in its pathophysiology. However, macrophage death during antibacterial activities will exacerbate inflammation and tissue damage. Porphyromonas gingivalis is a major constituent of subgingival biofilm plaques in periodontitis, but the effects and precise molecular mechanisms by which it triggers macrophage death remain unknown. Here we found that P. gingivalis infection notably activated multiple death pathways in bone-marrow-derived macrophages, including pyroptosis, apoptosis and necrosis. Furthermore, using RNA sequencing, we identified that P. gingivalis infection markedly increased the expression of Z-DNA binding protein 1 (Zbp1) in bone-marrow-derived macrophages. Initially identified as an interferon-induced tumor-associated protein, Zbp1 serves as an upstream sensor that regulates cell death by activating PANoptosis. Mechanistically, P. gingivalis induced a mitochondrial stress response, prompting the release of mitochondrial DNA. This mitochondrial DNA then interacted with Zbp1, consequently augmenting its downstream PANoptosis signals. In addition, P. gingivalis stimulated macrophage Zbp1 expression through the Tlr2/4-JNK-Stat3/5 pathway, exacerbating macrophage death. Importantly, blocking the biosynthesis of endogenous Zbp1 by pharmacological delivery with microneedles improved the survival of P. gingivalis-infected macrophages and inhibited periodontal tissue destruction. These findings highlight Zbp1 as a potential therapeutic target for P. gingivalis-induced periodontitis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Porphyromonas gingivalis Infection Induces Amyloid-β Accumulation in Monocytes/Macrophages.J Alzheimers Dis. 2019;72(2):479-494. doi: 10.3233/JAD-190298. J Alzheimers Dis. 2019. PMID: 31594220
-
Liver X receptors contribute to periodontal pathogen-elicited inflammation and oral bone loss.Mol Oral Microbiol. 2015 Dec;30(6):438-50. doi: 10.1111/omi.12103. Epub 2015 Jun 9. Mol Oral Microbiol. 2015. PMID: 25946408 Free PMC article.
-
The Anti-Inflammatory Effect of SDF-1 Derived Peptide on Porphyromonas gingivalis Infection via Regulation of NLRP3 and AIM2 Inflammasome.Pathogens. 2024 Jun 4;13(6):474. doi: 10.3390/pathogens13060474. Pathogens. 2024. PMID: 38921772 Free PMC article.
-
Porphyromonas gingivalis: major periodontopathic pathogen overview.J Immunol Res. 2014;2014:476068. doi: 10.1155/2014/476068. Epub 2014 Mar 25. J Immunol Res. 2014. PMID: 24741603 Free PMC article. Review.
-
Porphyromonas gingivalis mediated periodontal disease and atherosclerosis: disparate diseases with commonalities in pathogenesis through TLRs.Curr Pharm Des. 2007;13(36):3665-75. doi: 10.2174/138161207783018554. Curr Pharm Des. 2007. PMID: 18220804 Review.
Cited by
-
Bioinformatics-based screening and validation of PANoptosis-related biomarkers in periodontitis.Front Cell Dev Biol. 2025 Jun 19;13:1619002. doi: 10.3389/fcell.2025.1619002. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40612110 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- 82170993/National Natural Science Foundation of China (National Science Foundation of China)
- 82303457/National Natural Science Foundation of China (National Science Foundation of China)
- 82201097/National Natural Science Foundation of China (National Science Foundation of China)
- 2023M741789/China Postdoctoral Science Foundation
- 2023M741788 to X.H./China Postdoctoral Science Foundation
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

