mTOR controls ependymal cell differentiation by targeting the alternative cell cycle and centrosomal proteins
- PMID: 40307619
- PMCID: PMC12187940
- DOI: 10.1038/s44319-025-00460-2
mTOR controls ependymal cell differentiation by targeting the alternative cell cycle and centrosomal proteins
Abstract
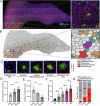
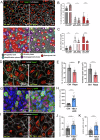
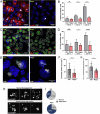
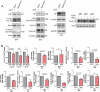
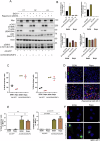
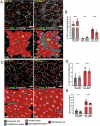
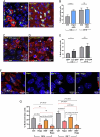
Ependymal cells are multiciliated glial cells lining the ventricles of the mammalian brain. Their differentiation from progenitor cells involves cell enlargement and progresses through centriole amplification phases and ciliogenesis. These phases are accompanied by the sharp up-regulation of mTOR Complex 1 activity (mTORC1), a master regulator of macromolecule biosynthesis and cell growth, whose function in ependymal cell differentiation is unknown. We demonstrate that mTORC1 inhibition by rapamycin preserves the progenitor pool by reinforcing quiescence and preventing alternative cell cycle progression for centriole amplification. Overexpressing E2F4 and MCIDAS circumvents mTORC1-regulated processes, enabling centriole amplification despite rapamycin, and enhancing mTORC1 activity through positive feedback. Acute rapamycin treatment in multicentriolar cells during the late phases of differentiation causes centriole regrouping, indicating a direct role of mTORC1 in centriole dynamics. By phosphoproteomic and phosphomutant analysis, we reveal that the mTORC1-mediated phosphorylation of GAS2L1, a centrosomal protein that links actin and microtubule cytoskeletons, participates in centriole disengagement. This multilayered and sequential control of ependymal development by mTORC1, from the progenitor pool to centriolar function, has implications for pathophysiological conditions like aging and hydrocephalus-prone genetic diseases.
Keywords: Cell Cycle; Ciliogenesis; Cytoskeleton; Differentiation; mTOR.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures






References
-
- Al Jord A, Shihavuddin A, Servignat d’Aout R, Faucourt M, Genovesio A, Karaiskou A, Sobczak-Thepot J, Spassky N, Meunier A (2017) Calibrated mitotic oscillator drives motile ciliogenesis. Science 358:803–806 - PubMed
-
- Al Jord A, Spassky N, Meunier A (2019) Motile ciliogenesis and the mitotic prism. Biol Cell 111:199–212 - PubMed
-
- Au FK, Jia Y, Jiang K, Grigoriev I, Hau BK, Shen Y, Du S, Akhmanova A, Qi RZ (2017) GAS2L1 is a centriole-associated protein required for centrosome dynamics and disjunction. Dev Cell 40:81–94 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

