Automated extraction of functional biomarkers of verbal and ambulatory ability from multi-institutional clinical notes using large language models
- PMID: 40307685
- PMCID: PMC12042395
- DOI: 10.1186/s11689-025-09612-w
Automated extraction of functional biomarkers of verbal and ambulatory ability from multi-institutional clinical notes using large language models
Abstract
Background: Functional biomarkers in neurodevelopmental disorders, such as verbal and ambulatory abilities, are essential for clinical care and research activities. Treatment planning, intervention monitoring, and identifying comorbid conditions in individuals with intellectual and developmental disabilities (IDDs) rely on standardized assessments of these abilities. However, traditional assessments impose a burden on patients and providers, often leading to longitudinal inconsistencies and inequities due to evolving guidelines and associated time-cost. Therefore, this study aimed to develop an automated approach to classify verbal and ambulatory abilities from EHR data of IDD and cerebral palsy (CP) patients. Application of large language models (LLMs) to clinical notes, which are rich in longitudinal data, may provide a low-burden pipeline for extracting functional biomarkers efficiently and accurately.
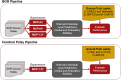
Methods: Data from the multi-institutional National Brain Gene Registry (BGR) and a CP clinic cohort were utilized, comprising 3,245 notes from 125 individuals and 5,462 clinical notes from 260 individuals, respectively. Employing three LLMs-GPT-3.5 Turbo, GPT-4 Turbo, and GPT-4 Omni-we provided the models with a clinical note and utilized a detailed conversational format to prompt the models to answer: "Does the individual use any words?" and "Can the individual walk without aid?" These responses were evaluated against ground-truth abilities, which were established using neurobehavioral assessments collected for each dataset.
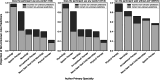
Results: LLM pipelines demonstrated high accuracy (weighted-F1 scores > .90) in predicting ambulatory ability for both cohorts, likely due to the consistent use of Gross Motor Functional Classification System (GMFCS) as a consistent ground-truth standard. However, verbal ability predictions were more accurate in the BGR cohort, likely due to higher adherence between the prompt and ground-truth assessment questions. While LLMs can be computationally expensive, analysis of our protocol affirmed the cost effectiveness when applied to select notes from the EHR.
Conclusions: LLMs are effective at extracting functional biomarkers from EHR data and broadly generalizable across variable note-taking practices and institutions. Individual verbal and ambulatory ability were accurately extracted, supporting the method's ability to streamline workflows by offering automated, efficient data extraction for patient care and research. Future studies are needed to extend this methodology to additional populations and to demonstrate more granular functional data classification.
Keywords: Electronic health records; Functional biomarkers; Large language models; Neurodevelopmental disorders.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: No participants were recruited specifically for this study. This work constitutes secondary use of data approved by the Washington University in St. Louis IRB (protocols #202010013 [Brain Gene Registry cohort] and #202309003 [cerebral palsy cohort]). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Biomarkers Definitions Working G. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69(3):89–95. 10.1067/mcp.2001.113989. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

