Emerging roles of exosomal circRNAs in non-small cell lung cancer
- PMID: 40307927
- PMCID: PMC12042431
- DOI: 10.1186/s12967-025-06463-w
Emerging roles of exosomal circRNAs in non-small cell lung cancer
Abstract
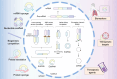
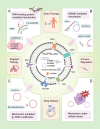
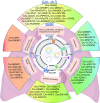
Despite the prevalence of non-small cell lung cancer (NSCLC) is high, the limited early detection and management of these tumors are restricted since there is an absence of reliable and precise diagnostic biomarkers and therapeutic targets. Exosomes transport functional molecules for facilitating intercellular communication, especially in the tumor microenvironment, indicating their potential as cancer biomarkers and therapeutic targets. Circular RNA (circRNA), a type of non-coding RNA possessing a covalently closed loop structure, substantial abundance, and tissue-specific expression patterns, is stably enriched in exosomes. In recent years, significant breakthroughs have been made in research on exosomal circRNA in NSCLC. This review briefly introduces the biogenesis, characterizations, and functions of circRNAs and exosomes, and systematically describes the biological functions and mechanisms of exosomal circRNAs in NSCLC. In addition, this study summarizes their role in the progression of NSCLC and discusses their clinical significance as biomarkers and therapeutic targets for NSCLC.
Keywords: Biomarker; CircRNAs; Exosomal circRNAs; Exosomes; Non-small cell lung cancer; Therapeutic target.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

