Analysis of the senescence secretome during zebrafish retina regeneration
- PMID: 40308558
- PMCID: PMC12040975
- DOI: 10.3389/fragi.2025.1569422
Analysis of the senescence secretome during zebrafish retina regeneration
Abstract
Introduction: Zebrafish possess the innate ability to regenerate any lost or damaged retinal cell type with Müller glia serving as resident stem cells. Recently, we discovered that this process is aided by a population of damage-induced senescent immune cells. As part of the Senescence Associated Secretory Phenotype (SASP), senescent cells secrete numerous factors that can play a role in the modulation of inflammation and remodeling of the retinal microenvironment during regeneration. However, the identity of specific SASP factors that drive initiation and progression of retina regeneration remains unclear.
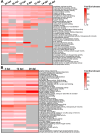
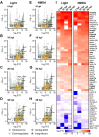
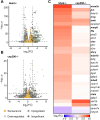
Materials and methods: We mined the SASP Atlas and publicly available RNAseq datasets to identify common, differentially expressed SASP factors after retina injury. These datasets included two distinct acute damage regimens, as well as two chronic, genetic models of retina degeneration. We identified overlapping factors between these models and used genetic knockdown experiments, qRT/PCR and immunohistochemical staining to test a role for one of these factors (npm1a).
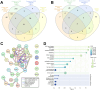
Results: We discovered an overlapping set of 31 SASP-related regeneration factors across all data sets and damage paradigms. These factors are upregulated after damage with functions that span the innate immune system, autophagic processing, cell cycle regulation, and cellular stress responses. From among these, we show that depletion of Nucleophosmin 1 (npm1a) inhibits retina regeneration and decreases senescent cell detection after damage.
Discussion: Our data suggest that differential expression of SASP factors promotes initiation and progression of retina regeneration after both acute and chronic retinal damage. The existence of a common, overlapping set of 31 factors provides a group of novel therapeutic targets for retina regeneration studies.
Keywords: SASP; inflammation; regeneration; retina; senescence; stemness.
Copyright © 2025 Konar, Vallone, Nguyen and Patton.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Damage-induced senescent immune cells regulate regeneration of the zebrafish retina.bioRxiv [Preprint]. 2023 Jan 16:2023.01.16.524296. doi: 10.1101/2023.01.16.524296. bioRxiv. 2023. Update in: Aging Biol. 2024;2:e20240021. doi: 10.59368/agingbio.20240021. PMID: 36711649 Free PMC article. Updated. Preprint.
-
Depletion of Polypyrimidine tract binding protein 1 (ptbp1) activates Müller glia-derived proliferation during zebrafish retina regeneration via modulation of the senescence secretome.Exp Eye Res. 2025 Aug;257:110420. doi: 10.1016/j.exer.2025.110420. Epub 2025 May 10. Exp Eye Res. 2025. PMID: 40355064
-
Dynamic changes in microglial and macrophage characteristics during degeneration and regeneration of the zebrafish retina.J Neuroinflammation. 2018 May 28;15(1):163. doi: 10.1186/s12974-018-1185-6. J Neuroinflammation. 2018. PMID: 29804544 Free PMC article.
-
Cellular senescence in the aging retina and developments of senotherapies for age-related macular degeneration.J Neuroinflammation. 2021 Jan 22;18(1):32. doi: 10.1186/s12974-021-02088-0. J Neuroinflammation. 2021. PMID: 33482879 Free PMC article. Review.
-
Potential Regulators of the Senescence-Associated Secretory Phenotype During Senescence and Aging.J Gerontol A Biol Sci Med Sci. 2022 Nov 21;77(11):2207-2218. doi: 10.1093/gerona/glac097. J Gerontol A Biol Sci Med Sci. 2022. PMID: 35524726 Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

